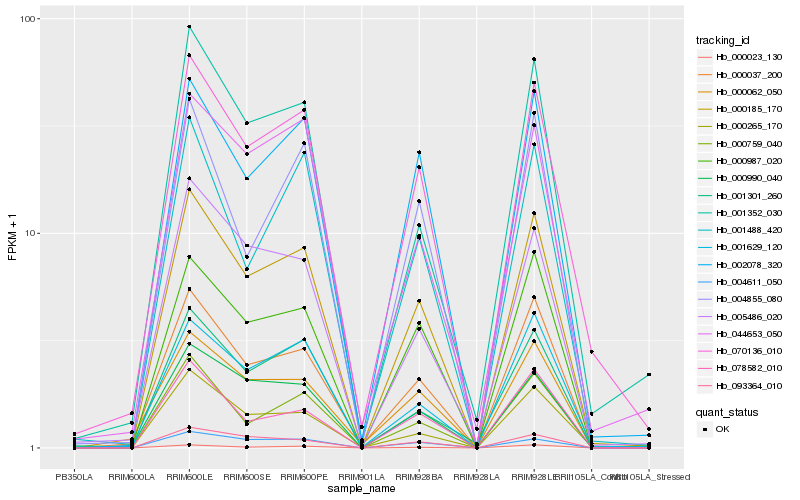
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000185_170 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_070136_010 |
0.0637706596 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 3 |
Hb_000037_200 |
0.0693714087 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_002078_320 |
0.0763666045 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_000987_020 |
0.0815109884 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 6 |
Hb_001629_120 |
0.089053074 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 7 |
Hb_000265_170 |
0.090712926 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 8 |
Hb_000023_130 |
0.0908870833 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 9 |
Hb_000759_040 |
0.0958148159 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 10 |
Hb_000990_040 |
0.0973225489 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 11 |
Hb_001352_030 |
0.0995833443 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 12 |
Hb_093364_010 |
0.1003560197 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 13 |
Hb_004611_050 |
0.1006255661 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 14 |
Hb_000062_050 |
0.1023990458 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_001301_260 |
0.1027868902 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005486_020 |
0.1039306557 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 17 |
Hb_078582_010 |
0.1050639471 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 18 |
Hb_044653_050 |
0.108087175 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001488_420 |
0.1087956524 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 20 |
Hb_004855_080 |
0.1109904146 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |