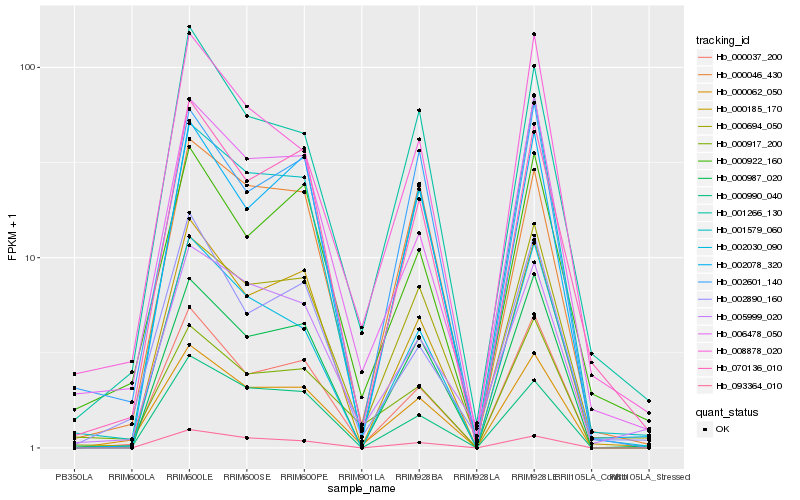
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_050 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000037_200 |
0.0768243977 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000185_170 |
0.1023990458 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000987_020 |
0.1067412377 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 5 |
Hb_070136_010 |
0.1127956499 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 6 |
Hb_001266_130 |
0.1141414442 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 7 |
Hb_000694_050 |
0.1159025398 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 8 |
Hb_006478_050 |
0.116355128 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_093364_010 |
0.1195249587 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 10 |
Hb_002601_140 |
0.1195600428 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 11 |
Hb_001579_060 |
0.120522468 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 12 |
Hb_000917_200 |
0.1210349394 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 13 |
Hb_008878_020 |
0.1213783613 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 14 |
Hb_000990_040 |
0.1218898739 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 15 |
Hb_000046_430 |
0.1232164522 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 16 |
Hb_002078_320 |
0.123607765 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_000922_160 |
0.1243272514 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 18 |
Hb_002890_160 |
0.1253824288 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002030_090 |
0.1270410992 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_005999_020 |
0.1306563961 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |