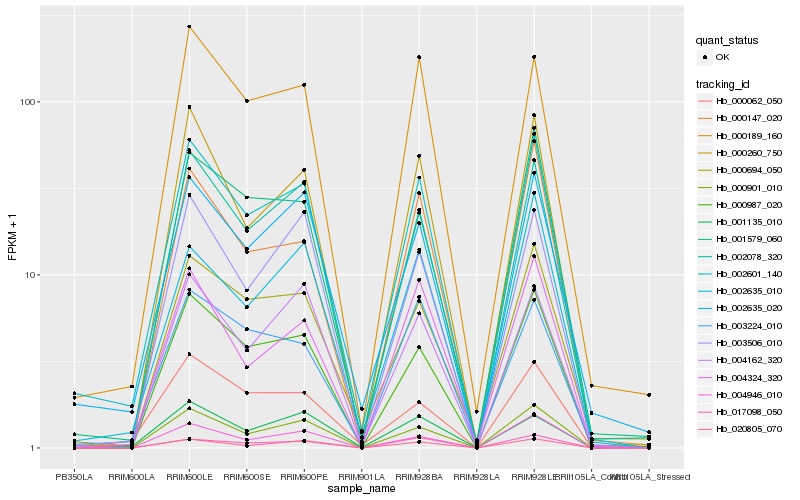
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002601_140 |
0.0 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 2 |
Hb_002635_020 |
0.0888718226 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 3 |
Hb_000260_750 |
0.0918131552 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 4 |
Hb_002078_320 |
0.0937521725 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_000987_020 |
0.0957735683 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 6 |
Hb_000694_050 |
0.0996247773 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 7 |
Hb_000901_010 |
0.1067076028 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 8 |
Hb_000189_160 |
0.1069748163 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 9 |
Hb_001579_060 |
0.1077362711 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 10 |
Hb_020805_070 |
0.112800398 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 11 |
Hb_003506_010 |
0.1169851128 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 12 |
Hb_004946_010 |
0.1186429796 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 13 |
Hb_004162_320 |
0.1187235089 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004324_320 |
0.1191096677 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 15 |
Hb_000147_020 |
0.1191534792 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 16 |
Hb_000062_050 |
0.1195600428 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_017098_050 |
0.1199206712 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 18 |
Hb_003224_010 |
0.1213428851 |
- |
- |
- |
| 19 |
Hb_001135_010 |
0.121355896 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 20 |
Hb_002635_010 |
0.1219909964 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |