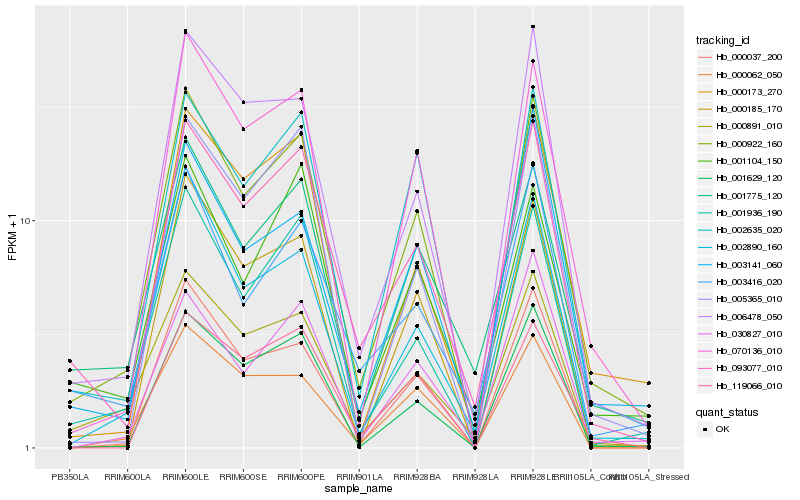
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_160 |
0.0 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 2 |
Hb_070136_010 |
0.0896212829 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 3 |
Hb_002635_020 |
0.0924896283 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 4 |
Hb_001936_190 |
0.0930901779 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 5 |
Hb_006478_050 |
0.098742055 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002890_160 |
0.0993321892 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_093077_010 |
0.0999097834 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000891_010 |
0.1048630435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000173_270 |
0.109012867 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001629_120 |
0.1097609672 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 11 |
Hb_001775_120 |
0.1105065265 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 12 |
Hb_000037_200 |
0.1182685238 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_003141_060 |
0.1188492917 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 14 |
Hb_003416_020 |
0.1191215681 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 15 |
Hb_001104_150 |
0.1193701745 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_000185_170 |
0.1194428479 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_005365_010 |
0.1196694513 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 18 |
Hb_030827_010 |
0.1242021571 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 19 |
Hb_000062_050 |
0.1243272514 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_119066_010 |
0.1250473649 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |