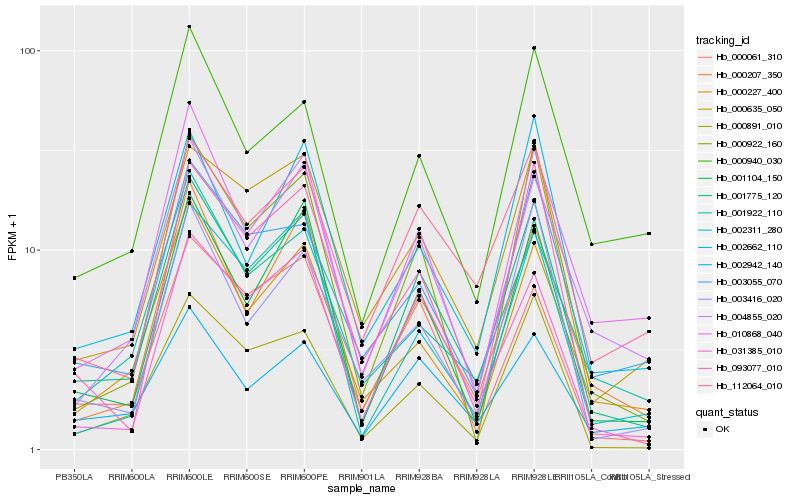
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_120 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 2 |
Hb_001104_150 |
0.0925235892 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 3 |
Hb_000207_350 |
0.1053601236 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 4 |
Hb_000922_160 |
0.1105065265 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 5 |
Hb_000891_010 |
0.111618853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_031385_010 |
0.1121483954 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 7 |
Hb_004855_020 |
0.1149184688 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 8 |
Hb_010868_040 |
0.1191347579 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 9 |
Hb_000227_400 |
0.1215915889 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 10 |
Hb_000635_050 |
0.1220032706 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 11 |
Hb_112064_010 |
0.1251314688 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 12 |
Hb_000061_310 |
0.1253112287 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002942_140 |
0.1255989531 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 14 |
Hb_093077_010 |
0.1258051568 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002311_280 |
0.1298138113 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_002662_110 |
0.1301579837 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 17 |
Hb_003055_070 |
0.1336020352 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000940_030 |
0.134876343 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003416_020 |
0.1352717379 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 20 |
Hb_001922_110 |
0.1362302384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |