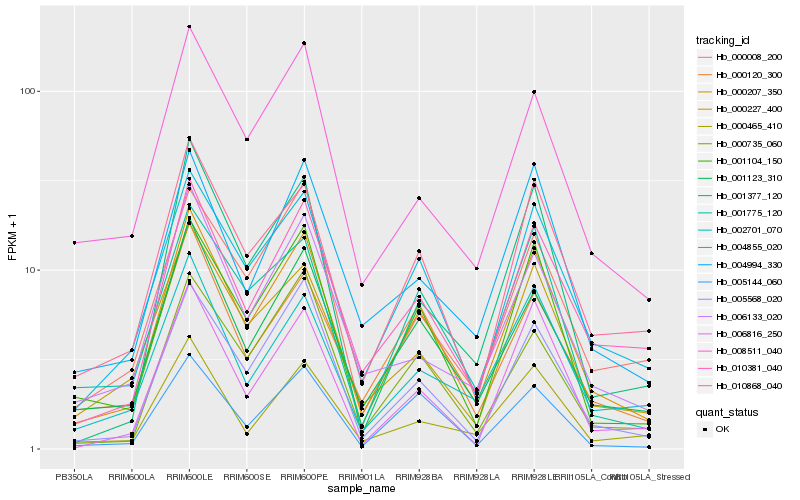
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_350 |
0.0 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 2 |
Hb_004855_020 |
0.075026316 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 3 |
Hb_001104_150 |
0.083753189 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 4 |
Hb_001123_310 |
0.0871663772 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 5 |
Hb_010868_040 |
0.0894804228 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 6 |
Hb_010381_040 |
0.0916611777 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 7 |
Hb_005568_020 |
0.0965806679 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000227_400 |
0.1017053252 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 9 |
Hb_004994_330 |
0.105078976 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001775_120 |
0.1053601236 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 11 |
Hb_002701_070 |
0.1106808668 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 12 |
Hb_005144_060 |
0.1150516051 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 13 |
Hb_000120_300 |
0.1153948257 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 14 |
Hb_001377_120 |
0.1171900854 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 15 |
Hb_008511_040 |
0.1184967793 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_000465_410 |
0.1186367693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000735_060 |
0.118897924 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 18 |
Hb_000008_200 |
0.1195467998 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 19 |
Hb_006816_250 |
0.119678161 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 20 |
Hb_006133_020 |
0.1211524684 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |