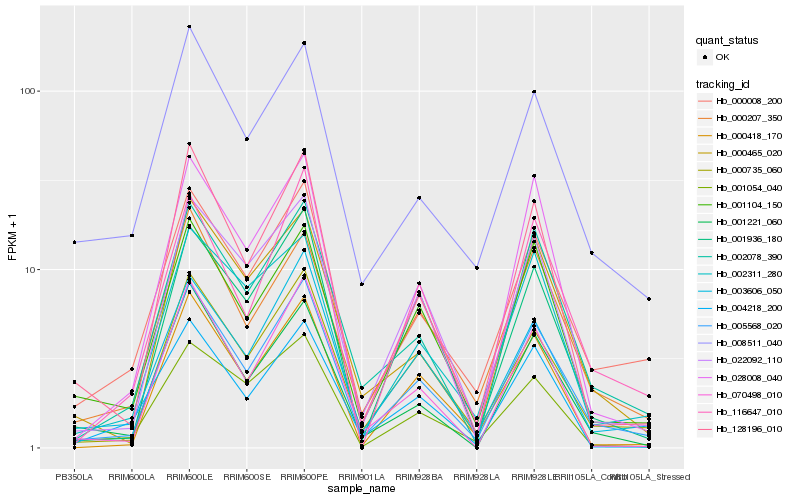
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005568_020 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000735_060 |
0.0769424752 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 3 |
Hb_002078_390 |
0.08363381 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 4 |
Hb_003606_050 |
0.0857378272 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 5 |
Hb_000008_200 |
0.0882230624 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 6 |
Hb_128196_010 |
0.0934953485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000207_350 |
0.0965806679 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 8 |
Hb_022092_110 |
0.0981511648 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 9 |
Hb_001104_150 |
0.0997748848 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 10 |
Hb_116647_010 |
0.1024596654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_028008_040 |
0.105848467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000418_170 |
0.1065887278 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 13 |
Hb_001221_060 |
0.1074946586 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 14 |
Hb_070498_010 |
0.1082059858 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 15 |
Hb_000465_020 |
0.1095709785 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 16 |
Hb_001936_180 |
0.1102287764 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 17 |
Hb_002311_280 |
0.1131115528 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_008511_040 |
0.1195464744 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 19 |
Hb_001054_040 |
0.1200885004 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 20 |
Hb_004218_200 |
0.12209477 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |