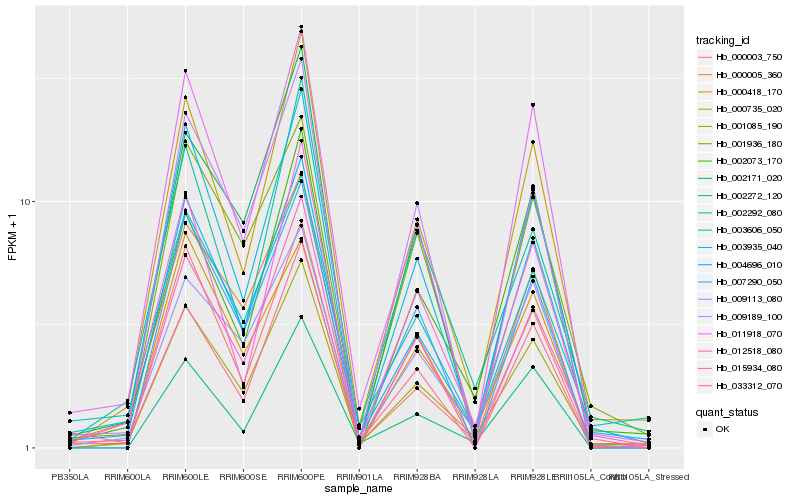
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_190 |
0.0 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_000735_020 |
0.053583102 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 3 |
Hb_011918_070 |
0.092075083 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 4 |
Hb_009113_080 |
0.0996282442 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 5 |
Hb_012518_080 |
0.1004573526 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 6 |
Hb_000005_360 |
0.1049447813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003606_050 |
0.1053434348 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 8 |
Hb_033312_070 |
0.1059659482 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 9 |
Hb_000003_750 |
0.1059826363 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 10 |
Hb_001936_180 |
0.1077058479 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 11 |
Hb_007290_050 |
0.1084380463 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 12 |
Hb_015934_080 |
0.1094699113 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 13 |
Hb_002292_080 |
0.1095735209 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 14 |
Hb_009189_100 |
0.1099966985 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 15 |
Hb_003935_040 |
0.1102431553 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 16 |
Hb_002272_120 |
0.1116761913 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 17 |
Hb_000418_170 |
0.1130315923 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 18 |
Hb_002171_020 |
0.1157037861 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 19 |
Hb_002073_170 |
0.1179758982 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 20 |
Hb_004696_010 |
0.1196129833 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |