| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
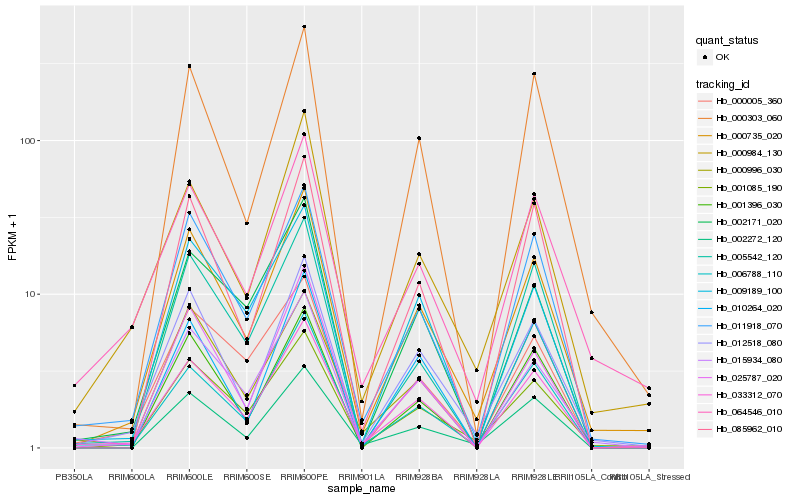
Hb_033312_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 2 |
Hb_006788_110 |
0.071345603 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 3 |
Hb_000735_020 |
0.083991002 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_011918_070 |
0.0860321072 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 5 |
Hb_015934_080 |
0.0909378728 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 6 |
Hb_012518_080 |
0.1050716838 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 7 |
Hb_001085_190 |
0.1059659482 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_025787_020 |
0.1064112014 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_002171_020 |
0.1083104948 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_000996_030 |
0.1088935047 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009189_100 |
0.1131864276 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 12 |
Hb_000303_060 |
0.1139839413 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_085962_010 |
0.1143092501 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 14 |
Hb_000005_360 |
0.1177115113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_064546_010 |
0.1178105861 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 16 |
Hb_005542_120 |
0.1184419469 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 17 |
Hb_000984_130 |
0.1195411366 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 18 |
Hb_002272_120 |
0.120810899 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 19 |
Hb_001396_030 |
0.120983237 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 20 |
Hb_010264_020 |
0.1219031276 |
- |
- |
unnamed protein product [Vitis vinifera] |