| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
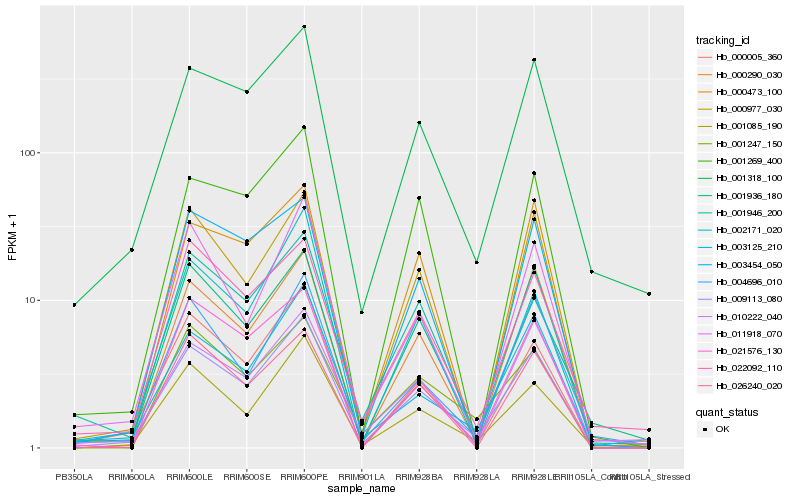
Hb_009113_080 |
0.0 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 2 |
Hb_001946_200 |
0.0941957557 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_001085_190 |
0.0996282442 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_000977_030 |
0.1013508908 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000005_360 |
0.1016142087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003125_210 |
0.1027588532 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 7 |
Hb_001318_100 |
0.104177993 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 8 |
Hb_000290_030 |
0.1049844484 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 9 |
Hb_011918_070 |
0.1134070972 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 10 |
Hb_004696_010 |
0.1157912878 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 11 |
Hb_026240_020 |
0.1184375661 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 12 |
Hb_001936_180 |
0.118913845 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 13 |
Hb_001269_400 |
0.1189662377 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 14 |
Hb_001247_150 |
0.1202369886 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 15 |
Hb_010222_040 |
0.1222608147 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 16 |
Hb_003454_050 |
0.1226422229 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 17 |
Hb_000473_100 |
0.1243731005 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 18 |
Hb_022092_110 |
0.1253261061 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 19 |
Hb_021576_130 |
0.1271418568 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002171_020 |
0.1281018351 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |