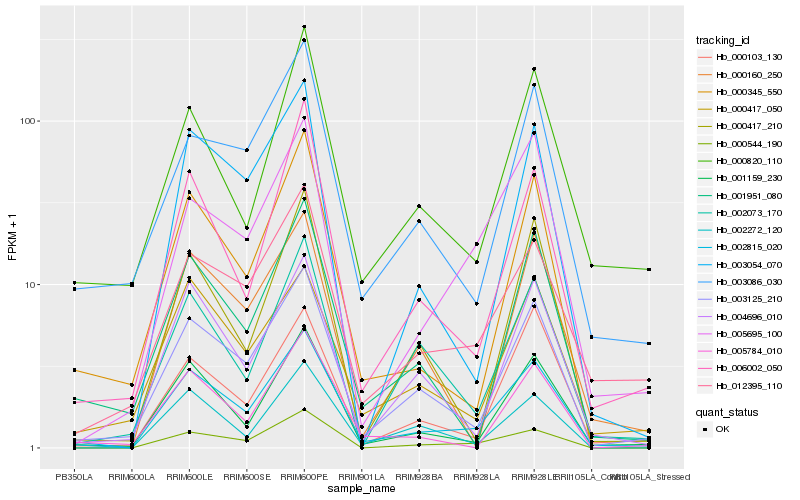
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002815_020 |
0.0 |
transcription factor |
TF Family: G2-like |
Homeodomain-like superfamily protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_003125_210 |
0.1039646853 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 3 |
Hb_001159_230 |
0.1088944785 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000544_190 |
0.1211371055 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 5 |
Hb_006002_050 |
0.134419305 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 6 |
Hb_004696_010 |
0.1358775663 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 7 |
Hb_012395_110 |
0.1371389848 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 8 |
Hb_000345_550 |
0.1402913379 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 9 |
Hb_003054_070 |
0.1403072548 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_005695_100 |
0.1434576062 |
- |
- |
PREDICTED: uncharacterized protein LOC105638839 [Jatropha curcas] |
| 11 |
Hb_001951_080 |
0.1469797698 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000417_050 |
0.147161095 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 13 |
Hb_000103_130 |
0.1504549286 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_002073_170 |
0.153271996 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 15 |
Hb_003086_030 |
0.1534075426 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 16 |
Hb_005784_010 |
0.1546889016 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 17 |
Hb_000417_210 |
0.1555792516 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_002272_120 |
0.1556375848 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 19 |
Hb_000820_110 |
0.1560582491 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000160_250 |
0.1564033843 |
- |
- |
Uncharacterized protein TCM_019235 [Theobroma cacao] |