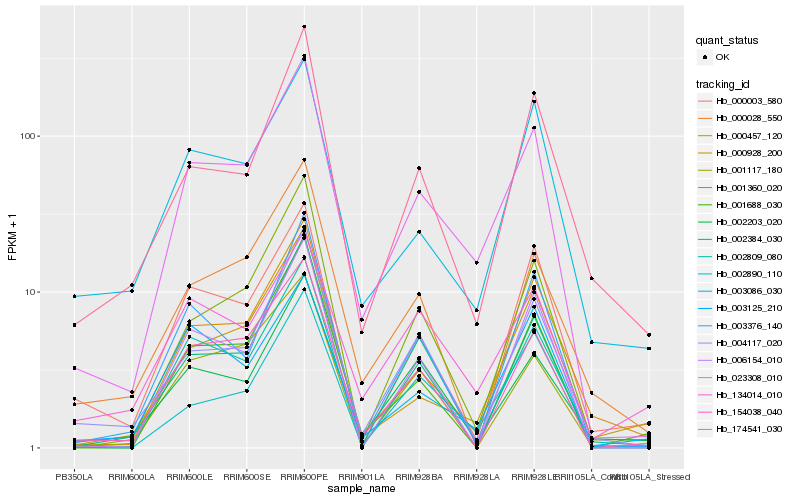
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023308_010 |
0.0 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 2 |
Hb_154038_040 |
0.1224713495 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 3 |
Hb_000457_120 |
0.1277403115 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 4 |
Hb_004117_020 |
0.1333273939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001360_020 |
0.133578058 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 6 |
Hb_000928_200 |
0.1409677263 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 7 |
Hb_001688_030 |
0.1411338722 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 8 |
Hb_002203_020 |
0.1420502001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003086_030 |
0.142953442 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 10 |
Hb_006154_010 |
0.1435563538 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_002809_080 |
0.1453420453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003376_140 |
0.1466566379 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 13 |
Hb_002384_030 |
0.1472970801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_174541_030 |
0.1472992397 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 15 |
Hb_002890_110 |
0.1485057148 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 16 |
Hb_003125_210 |
0.1495454631 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 17 |
Hb_000003_580 |
0.1513042248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_134014_010 |
0.1532066641 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_000028_550 |
0.1533603059 |
- |
- |
unknown [Medicago truncatula] |
| 20 |
Hb_001117_180 |
0.1539541641 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |