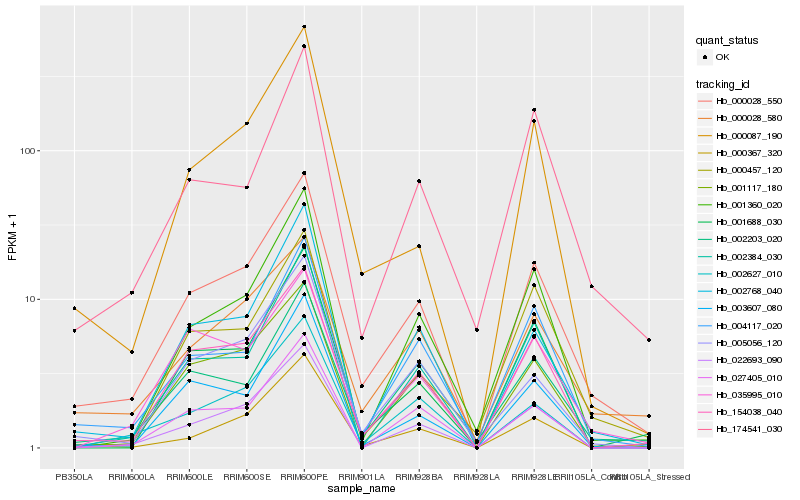
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_550 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_154038_040 |
0.1035061055 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 3 |
Hb_000028_580 |
0.1084477553 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 4 |
Hb_004117_020 |
0.1128284415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002768_040 |
0.1197842498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022693_090 |
0.1224010689 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001117_180 |
0.1232508524 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002203_020 |
0.1249582283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005056_120 |
0.1277692048 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000087_190 |
0.1319592495 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 11 |
Hb_001360_020 |
0.1328147074 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 12 |
Hb_002627_010 |
0.1377628697 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 13 |
Hb_035995_010 |
0.1378233081 |
- |
- |
- |
| 14 |
Hb_174541_030 |
0.1379572335 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 15 |
Hb_003607_080 |
0.1395910277 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 16 |
Hb_001688_030 |
0.1397131789 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 17 |
Hb_002384_030 |
0.1410541439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000367_320 |
0.1424141282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000457_120 |
0.1431411913 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 20 |
Hb_027405_010 |
0.14576459 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |