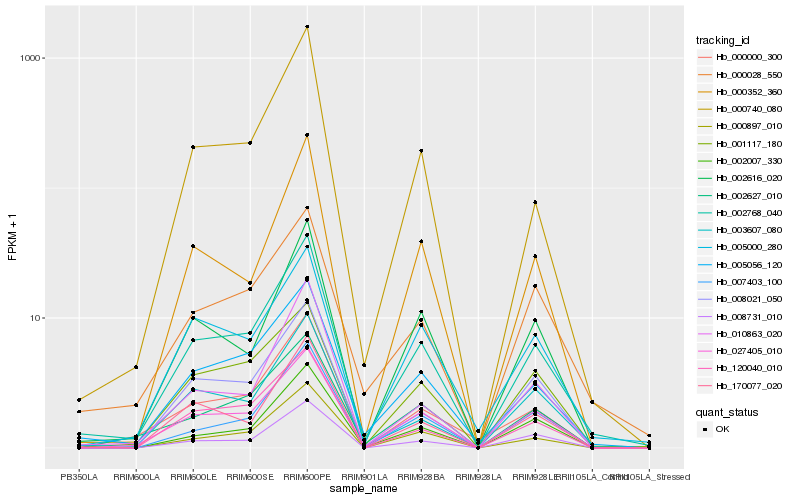
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002768_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005056_120 |
0.0841254474 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_027405_010 |
0.1026069627 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 4 |
Hb_003607_080 |
0.1057697931 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 5 |
Hb_000897_010 |
0.1095849288 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 6 |
Hb_000740_080 |
0.109889563 |
- |
- |
- |
| 7 |
Hb_000352_360 |
0.1157226798 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_002616_020 |
0.1172872185 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 9 |
Hb_000028_550 |
0.1197842498 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_120040_010 |
0.1197903398 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_010863_020 |
0.1198599467 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_170077_020 |
0.1202017827 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 13 |
Hb_007403_100 |
0.123905065 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 14 |
Hb_000000_300 |
0.1242078745 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 15 |
Hb_008731_010 |
0.1255340125 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 16 |
Hb_002627_010 |
0.1258737615 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 17 |
Hb_001117_180 |
0.1274639245 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005000_280 |
0.1276658291 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 19 |
Hb_002007_330 |
0.1278992319 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 20 |
Hb_008021_050 |
0.1279987308 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |