| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
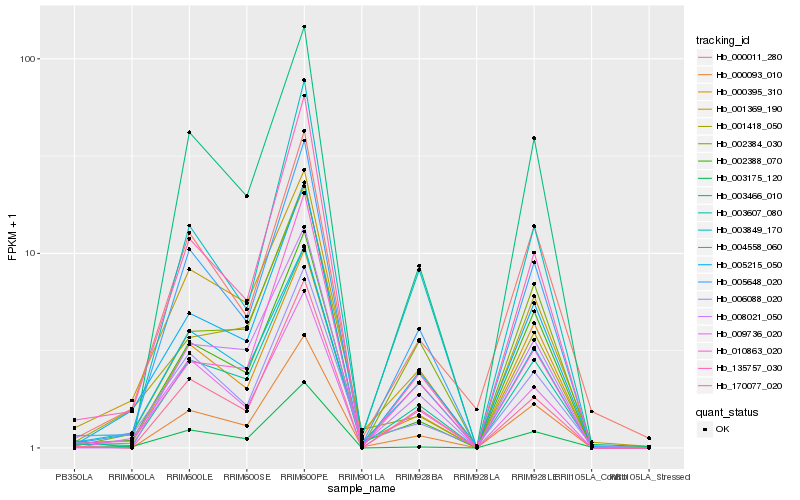
Hb_005215_050 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 2 |
Hb_003607_080 |
0.0801308341 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 3 |
Hb_005648_020 |
0.0882541119 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 4 |
Hb_002384_030 |
0.0963863965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001369_190 |
0.1019643147 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 6 |
Hb_004558_060 |
0.1044284274 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 7 |
Hb_008021_050 |
0.10833456 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_135757_030 |
0.1089829709 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 9 |
Hb_000093_010 |
0.1104264828 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 10 |
Hb_003849_170 |
0.1131913133 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 11 |
Hb_009736_020 |
0.1131976201 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 12 |
Hb_002388_070 |
0.1169116226 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_003466_010 |
0.1183840006 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 14 |
Hb_001418_050 |
0.11956264 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 15 |
Hb_006088_020 |
0.1205263661 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_170077_020 |
0.1224207644 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 17 |
Hb_000011_280 |
0.1228711903 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 18 |
Hb_010863_020 |
0.1234735746 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000395_310 |
0.1237778421 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 20 |
Hb_003175_120 |
0.124038312 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |