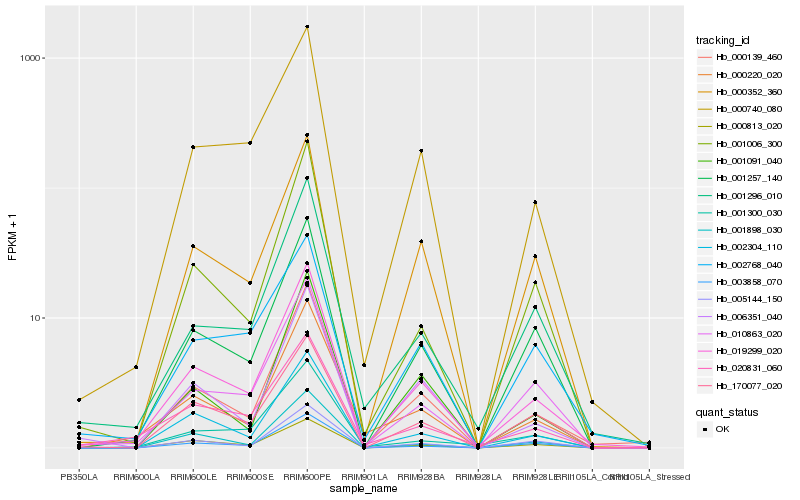
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019299_020 |
0.0 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 2 |
Hb_000139_460 |
0.073431817 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_000740_080 |
0.0903006926 |
- |
- |
- |
| 4 |
Hb_002304_110 |
0.0982018647 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 5 |
Hb_001257_140 |
0.1034285101 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 6 |
Hb_010863_020 |
0.1045426769 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000352_360 |
0.1055847954 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_000813_020 |
0.1095476673 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 9 |
Hb_000220_020 |
0.1117903769 |
- |
- |
hypothetical protein CICLE_v10017575mg [Citrus clementina] |
| 10 |
Hb_001296_010 |
0.1124241011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001006_300 |
0.1144433815 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_005144_150 |
0.1169169991 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 13 |
Hb_020831_060 |
0.1199888284 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 14 |
Hb_003858_070 |
0.120696323 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 15 |
Hb_170077_020 |
0.1209579068 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 16 |
Hb_006351_040 |
0.121972883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001898_030 |
0.1226216562 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 18 |
Hb_002768_040 |
0.130033708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001091_040 |
0.1301272576 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 20 |
Hb_001300_030 |
0.130439838 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |