| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
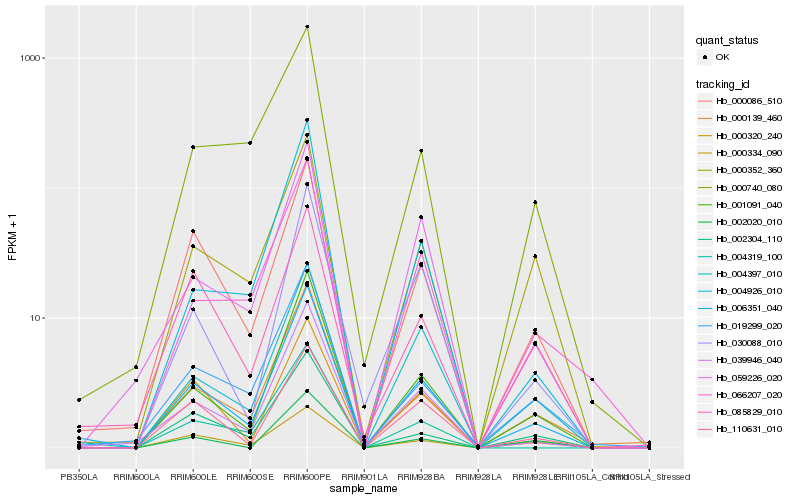
Hb_006351_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001091_040 |
0.0859945589 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 3 |
Hb_039946_040 |
0.1005306587 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 4 |
Hb_000139_460 |
0.1146843056 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 5 |
Hb_000086_510 |
0.121613167 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_019299_020 |
0.121972883 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 7 |
Hb_004397_010 |
0.1248302829 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_002304_110 |
0.1364621678 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 9 |
Hb_000320_240 |
0.1381201673 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_000352_360 |
0.1387435489 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 11 |
Hb_059226_020 |
0.1401232398 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 12 |
Hb_000334_090 |
0.1403349599 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 13 |
Hb_004926_010 |
0.1405666711 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 14 |
Hb_030088_010 |
0.1417600855 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 15 |
Hb_004319_100 |
0.1420235137 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 16 |
Hb_066207_020 |
0.1427830697 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 17 |
Hb_110631_010 |
0.1439002559 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 18 |
Hb_002020_010 |
0.1508069393 |
- |
- |
kinase-like protein [Corylus avellana] |
| 19 |
Hb_000740_080 |
0.1541786243 |
- |
- |
- |
| 20 |
Hb_085829_010 |
0.1543232815 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |