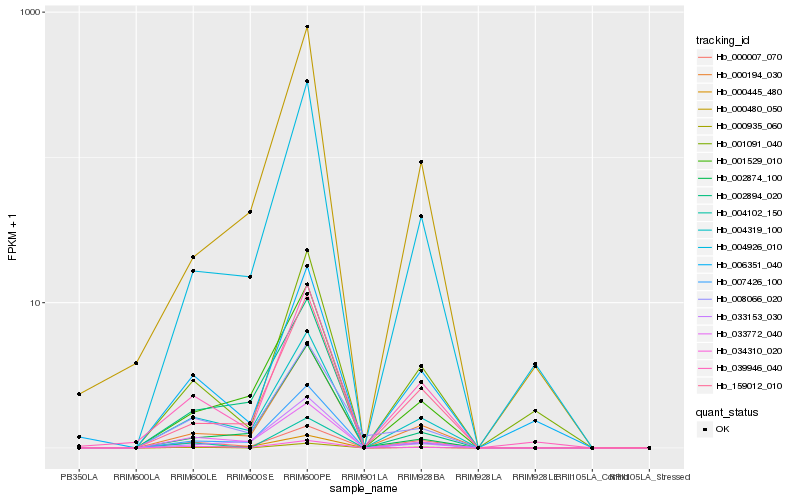
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004319_100 |
0.0 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000194_030 |
0.0680834272 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 3 |
Hb_033772_040 |
0.0791452836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007426_100 |
0.094549291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001529_010 |
0.0972313265 |
- |
- |
- |
| 6 |
Hb_034310_020 |
0.1010108189 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Jatropha curcas] |
| 7 |
Hb_159012_010 |
0.1050749521 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 8 |
Hb_004926_010 |
0.1114512473 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 9 |
Hb_039946_040 |
0.1119097214 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 10 |
Hb_033153_030 |
0.1332410493 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 11 |
Hb_000445_480 |
0.1402385802 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_006351_040 |
0.1420235137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008066_020 |
0.1421969607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002894_020 |
0.1455177414 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 15 |
Hb_000480_050 |
0.1474401708 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 16 |
Hb_002874_100 |
0.1501604703 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001091_040 |
0.1502371367 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 18 |
Hb_004102_150 |
0.1543142729 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 19 |
Hb_000007_070 |
0.1543876578 |
- |
- |
hypothetical protein POPTR_0008s11290g [Populus trichocarpa] |
| 20 |
Hb_000935_060 |
0.1551809101 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |