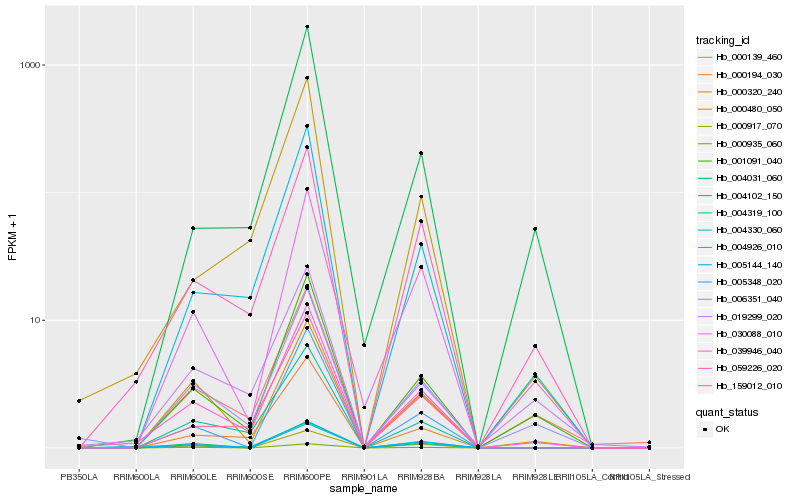
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039946_040 |
0.0 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 2 |
Hb_006351_040 |
0.1005306587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_059226_020 |
0.1088192719 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 4 |
Hb_001091_040 |
0.109056025 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 5 |
Hb_004319_100 |
0.1119097214 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 6 |
Hb_004926_010 |
0.1124979554 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 7 |
Hb_000194_030 |
0.1212737447 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 8 |
Hb_159012_010 |
0.1245730614 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 9 |
Hb_030088_010 |
0.1268199217 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 10 |
Hb_004330_060 |
0.1336740312 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 11 |
Hb_000139_460 |
0.1379948573 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 12 |
Hb_000917_070 |
0.138359594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004102_150 |
0.1385154776 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 14 |
Hb_000480_050 |
0.1404335196 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 15 |
Hb_005144_140 |
0.1420625999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000935_060 |
0.1421977386 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 17 |
Hb_019299_020 |
0.1462220103 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 18 |
Hb_005348_020 |
0.1473777696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000320_240 |
0.1478436855 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 20 |
Hb_004031_060 |
0.1501218636 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |