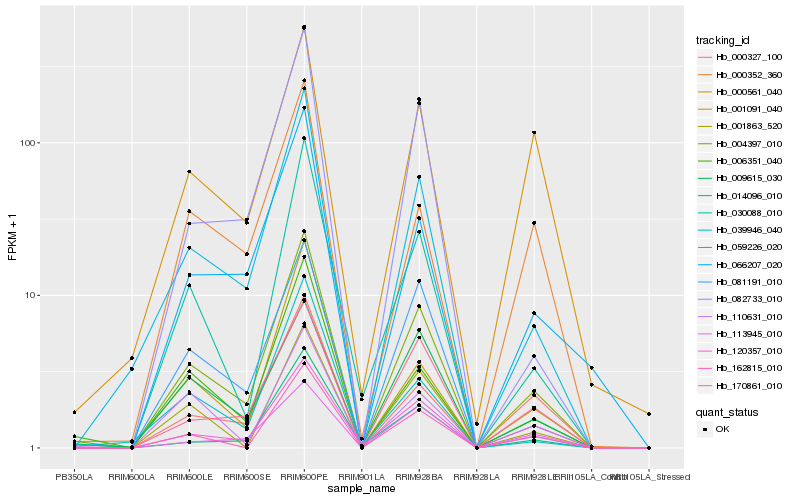
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004397_010 |
0.0 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 2 |
Hb_059226_020 |
0.0927669526 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 3 |
Hb_120357_010 |
0.1204148347 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_162815_010 |
0.1219903985 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_006351_040 |
0.1248302829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_030088_010 |
0.1251846687 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 7 |
Hb_066207_020 |
0.1298767621 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 8 |
Hb_014096_010 |
0.1343344455 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_082733_010 |
0.1354828873 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 10 |
Hb_081191_010 |
0.1356189634 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 11 |
Hb_170861_010 |
0.1368750813 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000327_100 |
0.1372643953 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 13 |
Hb_001863_520 |
0.1390397504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001091_040 |
0.1408822982 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 15 |
Hb_009615_030 |
0.1475277693 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000352_360 |
0.1495544441 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 17 |
Hb_000561_040 |
0.1512649099 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 18 |
Hb_113945_010 |
0.1517947267 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_110631_010 |
0.1522480011 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 20 |
Hb_039946_040 |
0.1527344674 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |