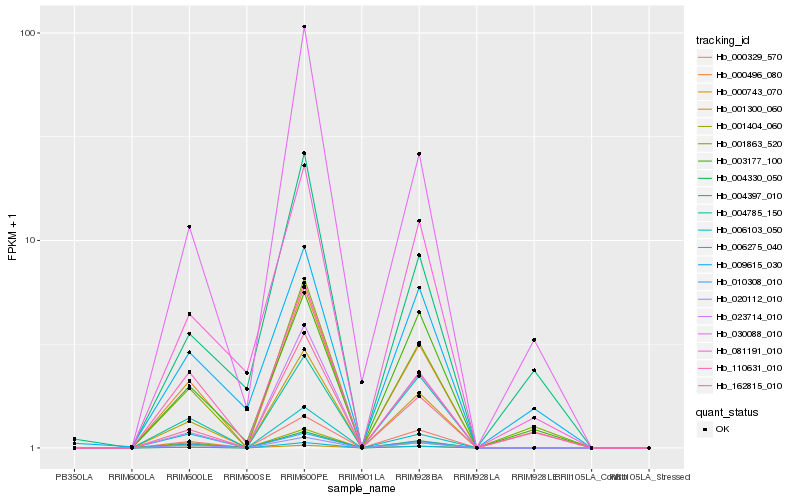
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_520 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_162815_010 |
0.0949240774 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_110631_010 |
0.1197262697 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 4 |
Hb_001300_060 |
0.1235241683 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 5 |
Hb_010308_010 |
0.1284267138 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 6 |
Hb_000496_080 |
0.128656917 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 7 |
Hb_004330_050 |
0.1291252148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_030088_010 |
0.1309205057 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 9 |
Hb_000329_570 |
0.1329786933 |
- |
- |
- |
| 10 |
Hb_020112_010 |
0.1335944662 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 11 |
Hb_006275_040 |
0.1347953513 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 12 |
Hb_003177_100 |
0.1373504218 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 13 |
Hb_004397_010 |
0.1390397504 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_081191_010 |
0.1529334812 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 15 |
Hb_009615_030 |
0.1582920013 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000743_070 |
0.1605513056 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 17 |
Hb_004785_150 |
0.1605614114 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 18 |
Hb_001404_060 |
0.1614216121 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_006103_050 |
0.1631100674 |
- |
- |
- |
| 20 |
Hb_023714_010 |
0.1632903391 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |