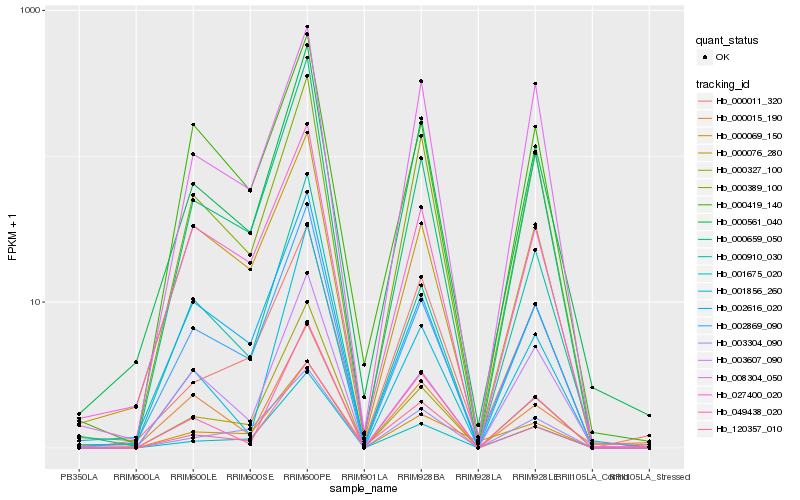
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000561_040 |
0.0 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 2 |
Hb_000389_100 |
0.0798675718 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 3 |
Hb_002869_090 |
0.0841323606 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 4 |
Hb_000659_050 |
0.0907560849 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 5 |
Hb_120357_010 |
0.0978485141 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_027400_020 |
0.1033763585 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 7 |
Hb_049438_020 |
0.1049615546 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 8 |
Hb_002616_020 |
0.1145783795 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 9 |
Hb_000011_320 |
0.1146442173 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 10 |
Hb_000327_100 |
0.1149818442 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 11 |
Hb_001675_020 |
0.1171755521 |
- |
- |
- |
| 12 |
Hb_000015_190 |
0.117610929 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 13 |
Hb_008304_050 |
0.1196788687 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 14 |
Hb_003304_090 |
0.1220217493 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 15 |
Hb_000419_140 |
0.1229536078 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 16 |
Hb_000076_280 |
0.127486651 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 17 |
Hb_000910_030 |
0.1276802045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003607_090 |
0.132580664 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 19 |
Hb_000069_150 |
0.1358455531 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 20 |
Hb_001856_260 |
0.1413509628 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |