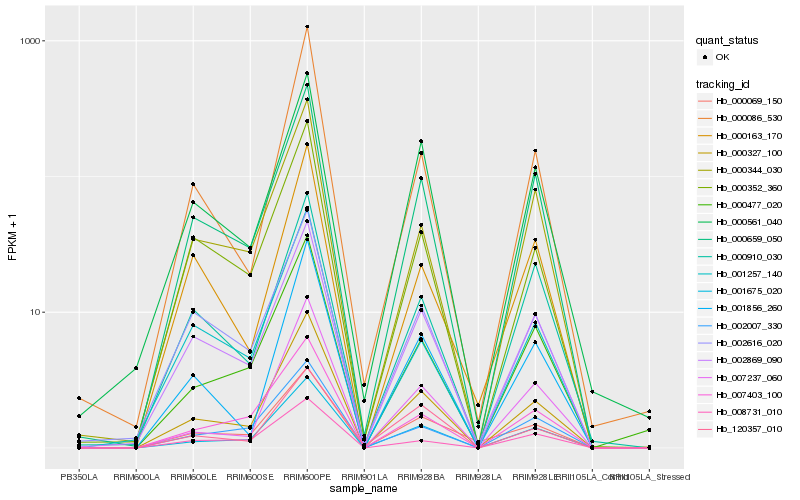
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 2 |
Hb_001675_020 |
0.0550596503 |
- |
- |
- |
| 3 |
Hb_000659_050 |
0.0736864115 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 4 |
Hb_002869_090 |
0.0784899898 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 5 |
Hb_000352_360 |
0.0876773898 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 6 |
Hb_001856_260 |
0.096146754 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 7 |
Hb_000477_020 |
0.0970267355 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 8 |
Hb_000086_530 |
0.0971922912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000344_030 |
0.0989341556 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 10 |
Hb_007237_060 |
0.1020595426 |
- |
- |
- |
| 11 |
Hb_007403_100 |
0.1023540203 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 12 |
Hb_002616_020 |
0.1046006882 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 13 |
Hb_001257_140 |
0.1116522512 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 14 |
Hb_002007_330 |
0.1148325154 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 15 |
Hb_000561_040 |
0.1149818442 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 16 |
Hb_000163_170 |
0.1256028449 |
- |
- |
- |
| 17 |
Hb_008731_010 |
0.1261078882 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 18 |
Hb_000069_150 |
0.1272020785 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 19 |
Hb_000910_030 |
0.1289797041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_120357_010 |
0.1294885035 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |