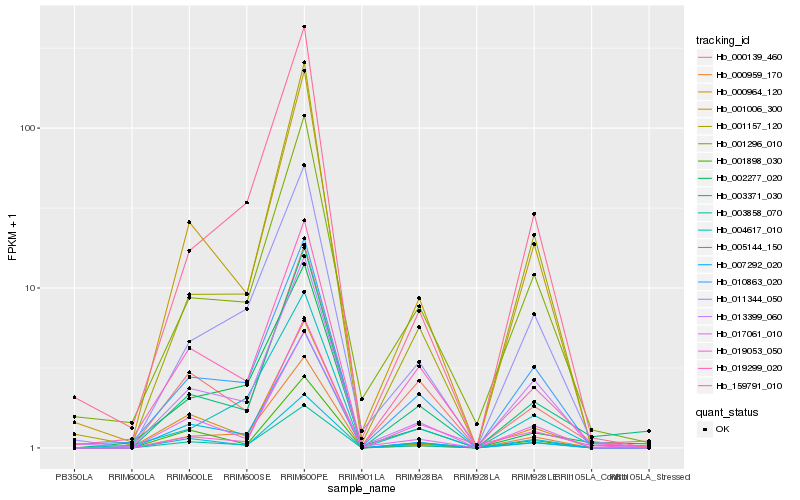
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_030 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000139_460 |
0.0991031776 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_001006_300 |
0.1291183049 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 4 |
Hb_003858_070 |
0.1302946233 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 5 |
Hb_019299_020 |
0.1334103334 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 6 |
Hb_001296_010 |
0.1343425885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_013399_060 |
0.138174605 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 8 |
Hb_019053_050 |
0.1394601834 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_005144_150 |
0.1432362842 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 10 |
Hb_004617_010 |
0.1434657753 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 11 |
Hb_017061_010 |
0.1443485457 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 12 |
Hb_001157_120 |
0.146770711 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 13 |
Hb_010863_020 |
0.1485542471 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000959_170 |
0.1491488567 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 15 |
Hb_007292_020 |
0.1497545907 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 16 |
Hb_001898_030 |
0.1498592346 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 17 |
Hb_011344_050 |
0.1523548723 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 18 |
Hb_002277_020 |
0.1530608838 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 19 |
Hb_159791_010 |
0.1536264913 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 20 |
Hb_000964_120 |
0.1556556298 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |