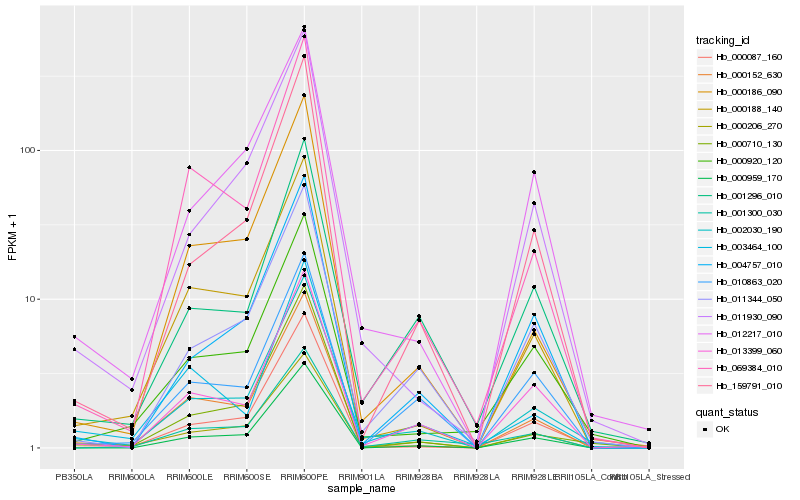
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000206_270 |
0.1019525629 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_011344_050 |
0.1042769358 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 4 |
Hb_001296_010 |
0.1057000609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000152_630 |
0.1057809665 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 6 |
Hb_000920_120 |
0.1102874085 |
- |
- |
- |
| 7 |
Hb_013399_060 |
0.1104661283 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 8 |
Hb_000188_140 |
0.1130758474 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 9 |
Hb_000087_160 |
0.1132703432 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_012217_010 |
0.1164937917 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 11 |
Hb_000186_090 |
0.1181384391 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 12 |
Hb_159791_010 |
0.1181749781 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 13 |
Hb_000959_170 |
0.1214960115 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 14 |
Hb_000710_130 |
0.1224867394 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_069384_010 |
0.1225575188 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_011930_090 |
0.1237137571 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 17 |
Hb_001300_030 |
0.1243429898 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |
| 18 |
Hb_004757_010 |
0.1253199927 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 19 |
Hb_010863_020 |
0.1265022245 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003464_100 |
0.127068216 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |