| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
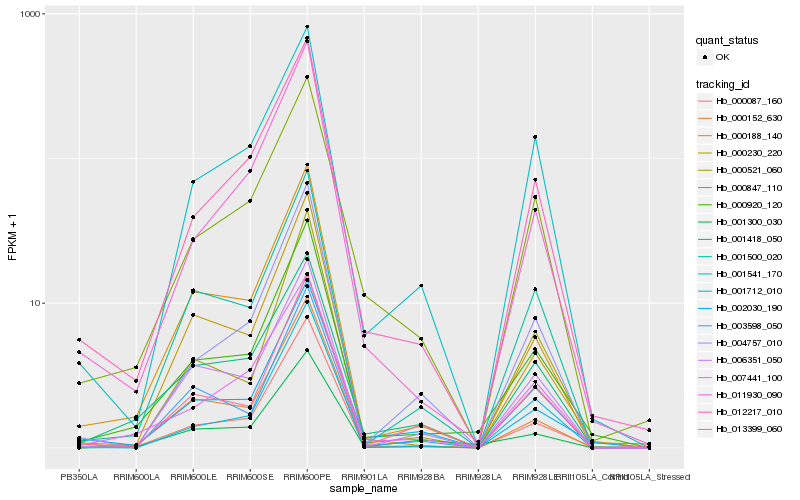
Hb_000920_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_000087_160 |
0.0939421051 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_012217_010 |
0.1060567075 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 4 |
Hb_002030_190 |
0.1102874085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007441_100 |
0.1104021495 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Populus euphratica] |
| 6 |
Hb_001500_020 |
0.1128157161 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000188_140 |
0.1149449604 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 8 |
Hb_000152_630 |
0.1156088453 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 9 |
Hb_001300_030 |
0.1156430639 |
- |
- |
hypothetical protein JCGZ_13871 [Jatropha curcas] |
| 10 |
Hb_001418_050 |
0.1162661989 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 11 |
Hb_006351_050 |
0.1184441965 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 12 |
Hb_000847_110 |
0.1190947607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011930_090 |
0.1199871559 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 14 |
Hb_001541_170 |
0.1205303934 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 15 |
Hb_000230_220 |
0.1233748098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003598_050 |
0.1239827587 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_013399_060 |
0.1240112846 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 18 |
Hb_000521_060 |
0.1251411448 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 19 |
Hb_004757_010 |
0.1268408354 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 20 |
Hb_001712_010 |
0.1271269833 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |