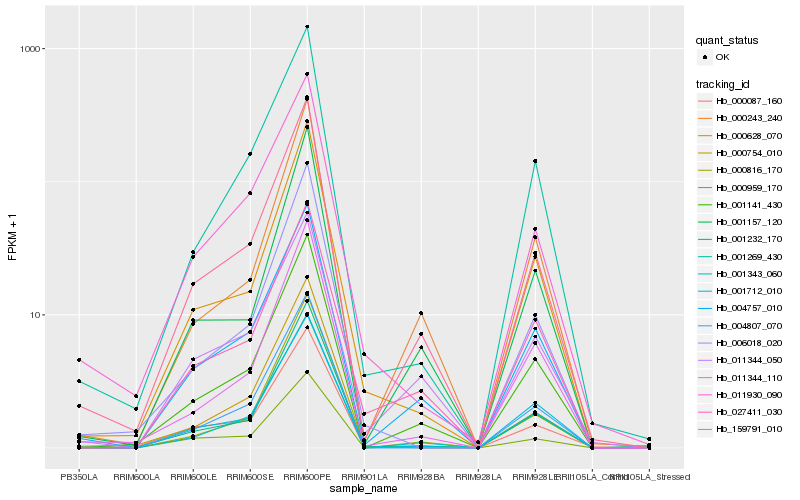
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159791_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 2 |
Hb_001141_430 |
0.0549166616 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 3 |
Hb_004757_010 |
0.0598248685 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 4 |
Hb_001343_060 |
0.0598403307 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 5 |
Hb_000959_170 |
0.0656804082 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 6 |
Hb_000816_170 |
0.0765488126 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 7 |
Hb_000087_160 |
0.0766154128 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_001232_170 |
0.0779888508 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 9 |
Hb_001157_120 |
0.0840655266 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 10 |
Hb_000243_240 |
0.086846259 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_000628_070 |
0.0899830614 |
- |
- |
- |
| 12 |
Hb_011344_110 |
0.0909541194 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 13 |
Hb_001269_430 |
0.0919092431 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 14 |
Hb_004807_070 |
0.0925991842 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 15 |
Hb_000754_010 |
0.0941835904 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001712_010 |
0.0951858932 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 17 |
Hb_011930_090 |
0.0961777148 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 18 |
Hb_006018_020 |
0.099200814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_027411_030 |
0.0999206922 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_011344_050 |
0.1008474504 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |