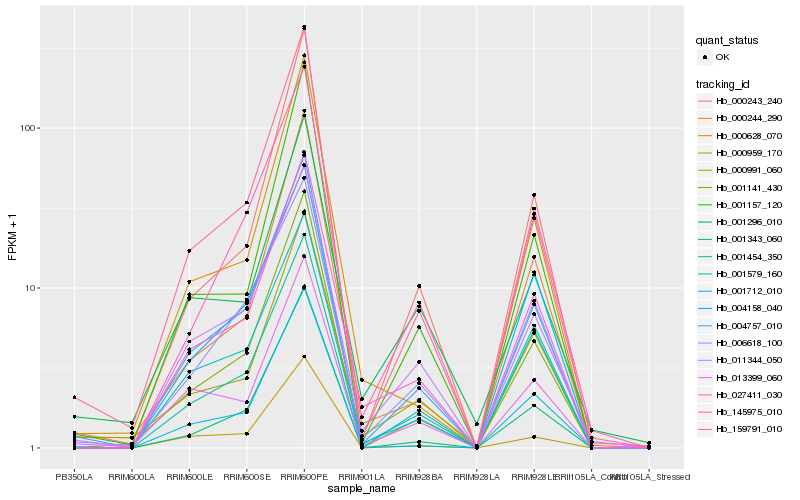
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027411_030 |
0.0 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_004757_010 |
0.0714654775 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 3 |
Hb_011344_050 |
0.0781045991 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 4 |
Hb_001579_160 |
0.0822038265 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 5 |
Hb_000991_060 |
0.0834304767 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 6 |
Hb_001141_430 |
0.085539706 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 7 |
Hb_145975_010 |
0.0915642642 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_001712_010 |
0.0949005384 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 9 |
Hb_000628_070 |
0.0958499602 |
- |
- |
- |
| 10 |
Hb_001343_060 |
0.0971547194 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 11 |
Hb_159791_010 |
0.0999206922 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 12 |
Hb_013399_060 |
0.1003406222 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 13 |
Hb_006618_100 |
0.1007697695 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 14 |
Hb_000244_290 |
0.1016733688 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000243_240 |
0.1064325297 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 16 |
Hb_000959_170 |
0.1080646174 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 17 |
Hb_004158_040 |
0.1093612928 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 18 |
Hb_001454_350 |
0.1095025039 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 19 |
Hb_001296_010 |
0.109833981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001157_120 |
0.1103251298 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |