| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
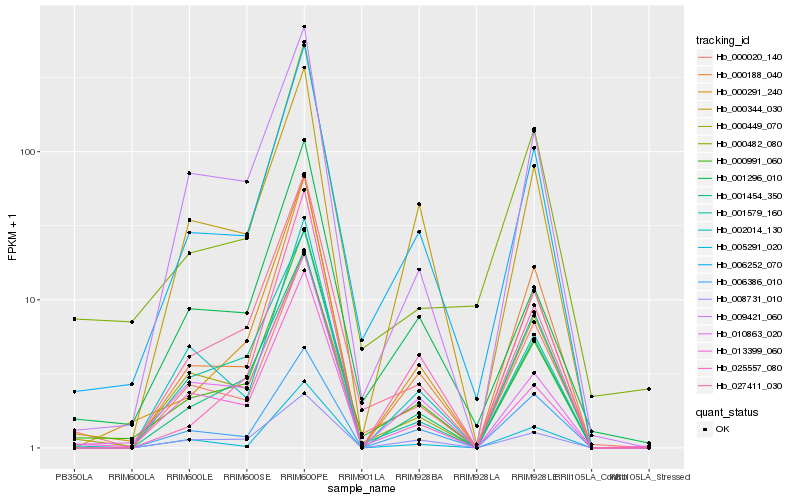
Hb_006252_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_17254 [Jatropha curcas] |
| 2 |
Hb_000991_060 |
0.070716868 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 3 |
Hb_000188_040 |
0.0943581408 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 4 |
Hb_000020_140 |
0.1051963967 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 5 |
Hb_001454_350 |
0.1056132948 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 6 |
Hb_002014_130 |
0.1091346644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000291_240 |
0.1098514711 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 8 |
Hb_000344_030 |
0.1106865468 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 9 |
Hb_001296_010 |
0.1113244092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_027411_030 |
0.1159047552 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_009421_060 |
0.1161553786 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 12 |
Hb_010863_020 |
0.1176845572 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_013399_060 |
0.1203749358 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 14 |
Hb_000449_070 |
0.1227341729 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_005291_020 |
0.1233657961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006386_010 |
0.1260856763 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 17 |
Hb_000482_080 |
0.1273262222 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 18 |
Hb_001579_160 |
0.1277054144 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 19 |
Hb_025557_080 |
0.1294230712 |
transcription factor |
TF Family: zf-HD |
- |
| 20 |
Hb_008731_010 |
0.1307981313 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |