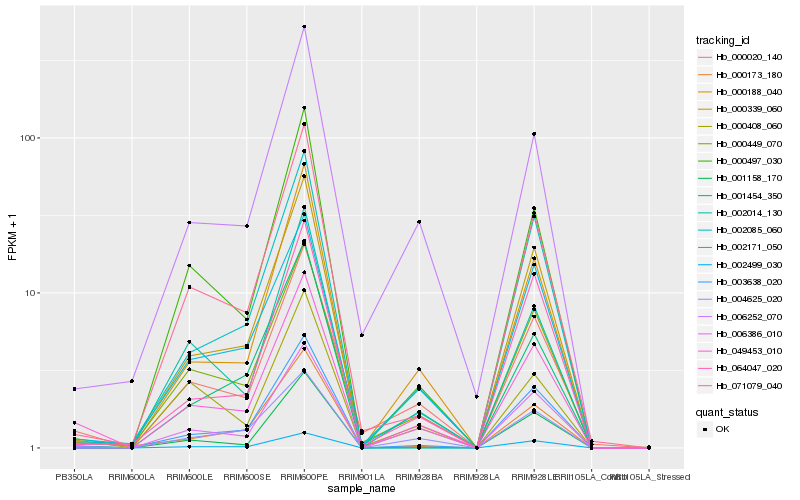
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049453_010 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000020_140 |
0.0918772281 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 3 |
Hb_000449_070 |
0.0984708309 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000173_180 |
0.1094431957 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004625_020 |
0.1195248376 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 6 |
Hb_000339_060 |
0.1265357184 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000188_040 |
0.1278563682 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 8 |
Hb_071079_040 |
0.1286872599 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 9 |
Hb_002171_050 |
0.1297695209 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 10 |
Hb_002014_130 |
0.1335524352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000408_060 |
0.134299261 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 12 |
Hb_003638_020 |
0.1355582425 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 13 |
Hb_006252_070 |
0.1366518113 |
- |
- |
hypothetical protein JCGZ_17254 [Jatropha curcas] |
| 14 |
Hb_001158_170 |
0.1369307167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002085_060 |
0.1372874905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001454_350 |
0.1376520602 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 17 |
Hb_000497_030 |
0.1383280742 |
- |
- |
- |
| 18 |
Hb_006386_010 |
0.1392514293 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 19 |
Hb_002499_030 |
0.140112045 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_064047_020 |
0.1419326648 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |