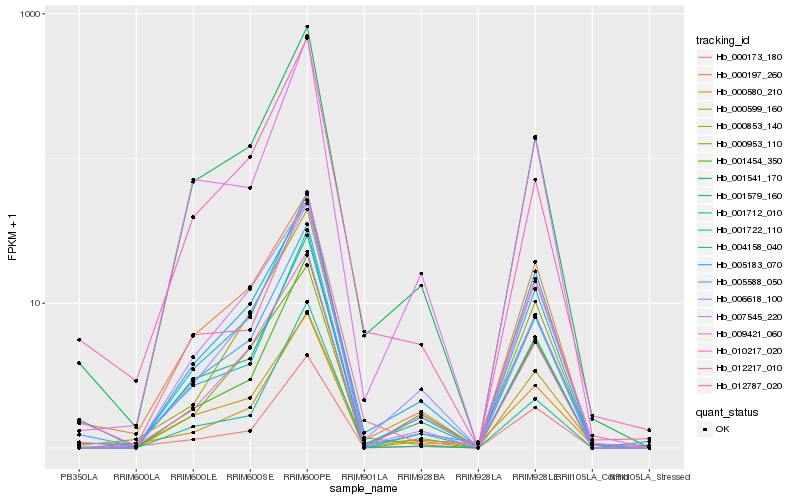
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004158_040 |
0.0659135796 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 3 |
Hb_001541_170 |
0.0753807623 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 4 |
Hb_001722_110 |
0.0836152978 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001579_160 |
0.086916134 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 6 |
Hb_012787_020 |
0.0926842596 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 7 |
Hb_000580_210 |
0.0940075411 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 8 |
Hb_005183_070 |
0.0948923355 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 9 |
Hb_007545_220 |
0.0953756616 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 10 |
Hb_000197_260 |
0.0980554864 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 11 |
Hb_010217_020 |
0.1022362642 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 12 |
Hb_000853_140 |
0.1026270443 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 13 |
Hb_006618_100 |
0.1029162798 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 14 |
Hb_009421_060 |
0.1031637679 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000173_180 |
0.10396415 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000953_110 |
0.1043500907 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 17 |
Hb_001454_350 |
0.1070616774 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 18 |
Hb_001712_010 |
0.1098326436 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 19 |
Hb_000599_160 |
0.1107782733 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 20 |
Hb_012217_010 |
0.1129954216 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |