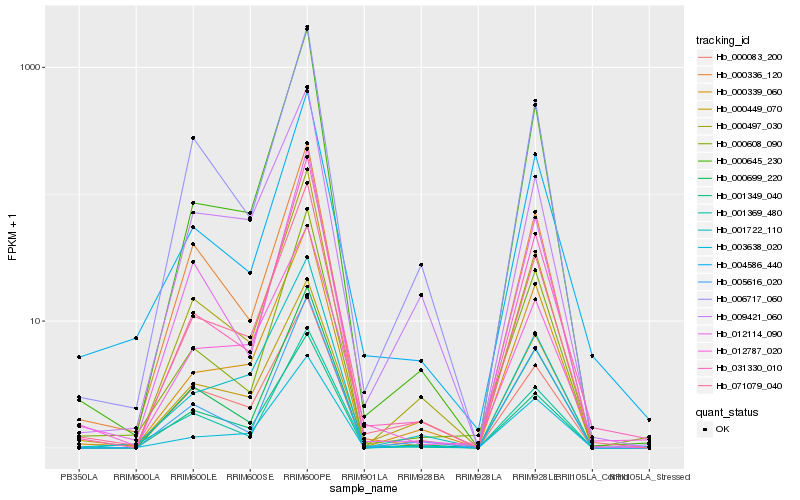
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071079_040 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 2 |
Hb_000497_030 |
0.0624361176 |
- |
- |
- |
| 3 |
Hb_000699_220 |
0.0695836948 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 4 |
Hb_000339_060 |
0.0696067509 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031330_010 |
0.0699612987 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000645_230 |
0.0728136925 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_006717_060 |
0.0776939161 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 8 |
Hb_001349_040 |
0.0785399655 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 9 |
Hb_001369_480 |
0.0812586283 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 10 |
Hb_000083_200 |
0.081669655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003638_020 |
0.0832527282 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 12 |
Hb_001722_110 |
0.0853859824 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_004586_440 |
0.0879157827 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 14 |
Hb_005616_020 |
0.0882883469 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_009421_060 |
0.0915043168 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 16 |
Hb_012114_090 |
0.0921295704 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 17 |
Hb_000608_090 |
0.0924838037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000449_070 |
0.0928563819 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_012787_020 |
0.0930542056 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 20 |
Hb_000336_120 |
0.093825111 |
- |
- |
PREDICTED: mavicyanin [Populus euphratica] |