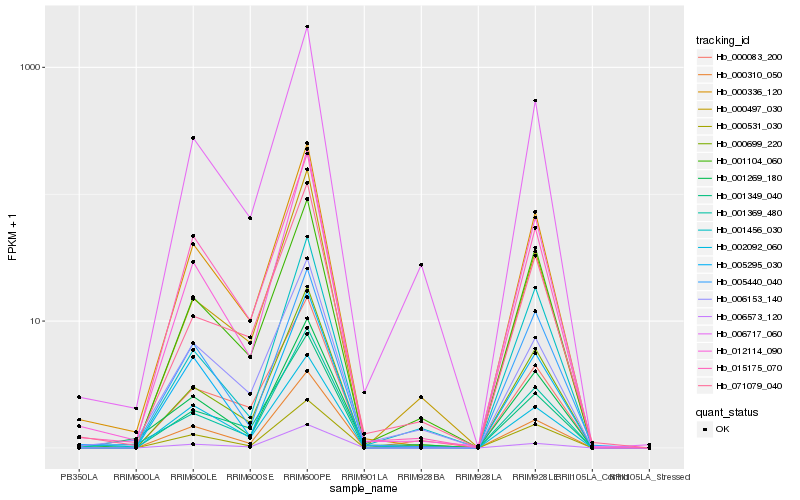
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_120 |
0.0 |
- |
- |
PREDICTED: mavicyanin [Populus euphratica] |
| 2 |
Hb_012114_090 |
0.0485661912 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 3 |
Hb_015175_070 |
0.058649963 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 4 |
Hb_000310_050 |
0.0596091808 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000699_220 |
0.0634124891 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 6 |
Hb_001369_480 |
0.0706788269 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 7 |
Hb_006717_060 |
0.0717306351 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 8 |
Hb_001349_040 |
0.0722103837 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 9 |
Hb_001104_060 |
0.0838919718 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 10 |
Hb_000531_030 |
0.0887385907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006573_120 |
0.0915224006 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |
| 12 |
Hb_002092_060 |
0.091731259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001269_180 |
0.0935874814 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_071079_040 |
0.093825111 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 15 |
Hb_000083_200 |
0.0962194757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006153_140 |
0.0973483684 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_005295_030 |
0.0982302731 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 18 |
Hb_005440_040 |
0.0982320705 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 19 |
Hb_000497_030 |
0.1004559092 |
- |
- |
- |
| 20 |
Hb_001456_030 |
0.1023881319 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |