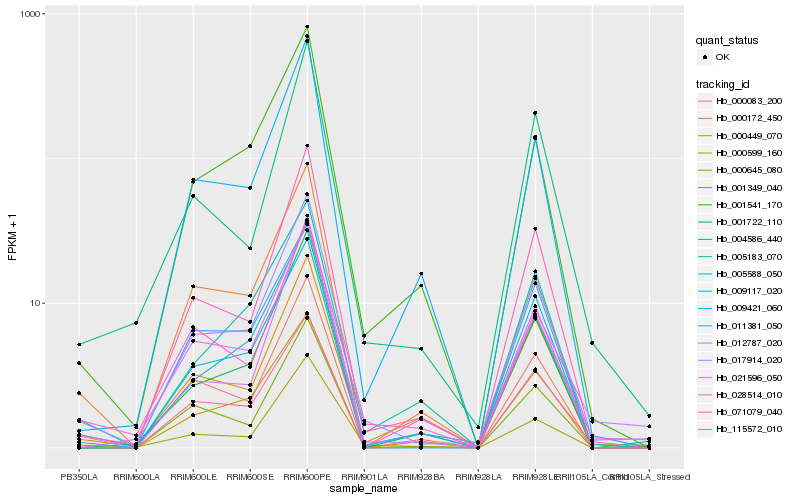
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012787_020 |
0.0 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 2 |
Hb_005588_050 |
0.0926842596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_071079_040 |
0.0930542056 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 4 |
Hb_000645_080 |
0.100770903 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_028514_010 |
0.1078429016 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 6 |
Hb_001541_170 |
0.1099099855 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 7 |
Hb_115572_010 |
0.1099849794 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 8 |
Hb_001722_110 |
0.1113172302 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_009421_060 |
0.1155619372 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_021596_050 |
0.1191795319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000449_070 |
0.1193673304 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_004586_440 |
0.1195523668 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 13 |
Hb_000083_200 |
0.1196932904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011381_050 |
0.120085964 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 15 |
Hb_017914_020 |
0.120119108 |
- |
- |
- |
| 16 |
Hb_001349_040 |
0.1202281546 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 17 |
Hb_009117_020 |
0.1227564696 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 18 |
Hb_000172_450 |
0.1232127994 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 19 |
Hb_005183_070 |
0.1236962255 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 20 |
Hb_000599_160 |
0.124088635 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |