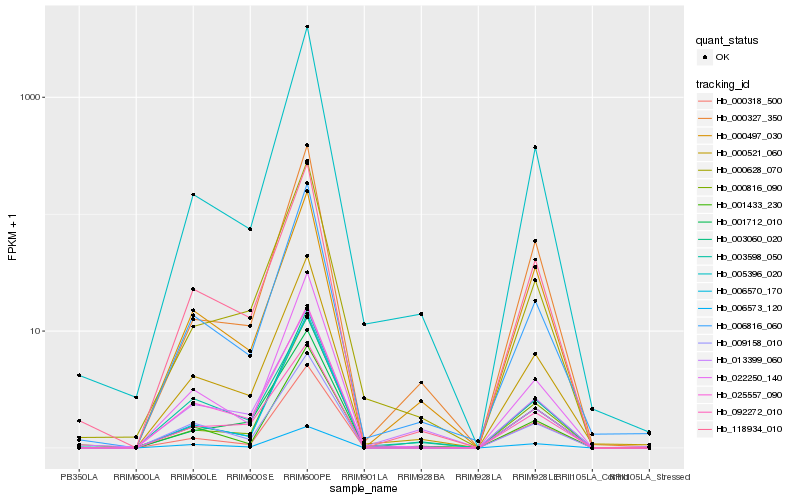
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_060 |
0.0 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 2 |
Hb_006816_060 |
0.0473527005 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 3 |
Hb_118934_010 |
0.0562256122 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_001712_010 |
0.0786070711 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 5 |
Hb_022250_140 |
0.0795548482 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_001433_230 |
0.084370041 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 7 |
Hb_003598_050 |
0.087279818 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000628_070 |
0.087440498 |
- |
- |
- |
| 9 |
Hb_009158_010 |
0.0882024605 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 10 |
Hb_000816_090 |
0.0883308107 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_005396_020 |
0.0894548228 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 12 |
Hb_025557_090 |
0.0937551588 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 13 |
Hb_092272_010 |
0.0941708743 |
- |
- |
- |
| 14 |
Hb_003060_020 |
0.0945954636 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_000327_350 |
0.0949758448 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 16 |
Hb_013399_060 |
0.0977420875 |
- |
- |
PREDICTED: uncharacterized protein LOC105133408 [Populus euphratica] |
| 17 |
Hb_006570_170 |
0.0996189553 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000497_030 |
0.0997779112 |
- |
- |
- |
| 19 |
Hb_000318_500 |
0.1001277715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006573_120 |
0.1017179155 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |