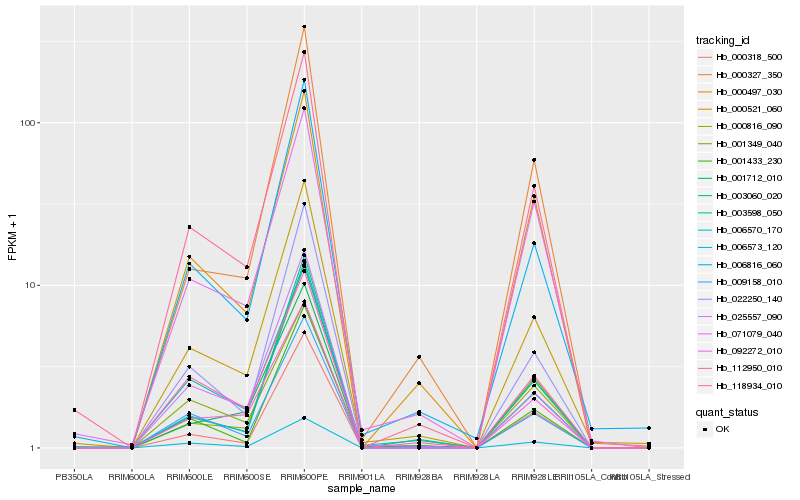
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118934_010 |
0.0 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_000521_060 |
0.0562256122 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 3 |
Hb_009158_010 |
0.0712914574 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 4 |
Hb_006573_120 |
0.0732715222 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |
| 5 |
Hb_092272_010 |
0.0735554659 |
- |
- |
- |
| 6 |
Hb_001712_010 |
0.0788708741 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_000497_030 |
0.080698557 |
- |
- |
- |
| 8 |
Hb_006816_060 |
0.0823118121 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 9 |
Hb_000318_500 |
0.0847125596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_022250_140 |
0.0865434905 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_003598_050 |
0.0865696859 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 12 |
Hb_025557_090 |
0.0907715954 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 13 |
Hb_000816_090 |
0.0920464798 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_112950_010 |
0.0923723961 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 15 |
Hb_071079_040 |
0.0958220573 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 16 |
Hb_001349_040 |
0.099645432 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 17 |
Hb_001433_230 |
0.100842937 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 18 |
Hb_006570_170 |
0.1014968928 |
- |
- |
Gibberellin-regulated protein 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000327_350 |
0.1016417956 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 20 |
Hb_003060_020 |
0.1049263594 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |