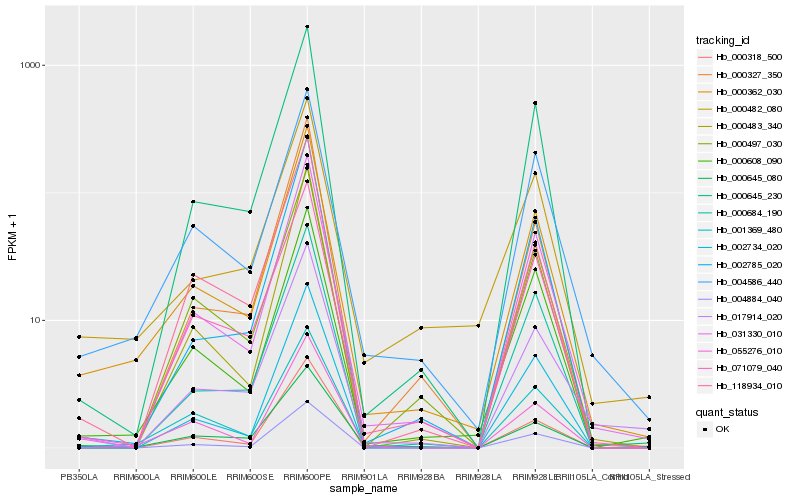
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_030 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 2 |
Hb_004586_440 |
0.0844230875 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_031330_010 |
0.0857057774 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001369_480 |
0.0913223012 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 5 |
Hb_000608_090 |
0.0923053195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000645_230 |
0.0960075921 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_000482_080 |
0.0962146288 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 8 |
Hb_071079_040 |
0.0986511682 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 9 |
Hb_000645_080 |
0.1062134532 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_002785_020 |
0.106275677 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 11 |
Hb_000483_340 |
0.1073661931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017914_020 |
0.1082387078 |
- |
- |
- |
| 13 |
Hb_055276_010 |
0.108414139 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 14 |
Hb_004884_040 |
0.1088585873 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 15 |
Hb_000684_190 |
0.1133623032 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000497_030 |
0.1157491535 |
- |
- |
- |
| 17 |
Hb_000318_500 |
0.1159499858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_118934_010 |
0.115989969 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_000327_350 |
0.116731279 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 20 |
Hb_002734_020 |
0.1167413544 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |