| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
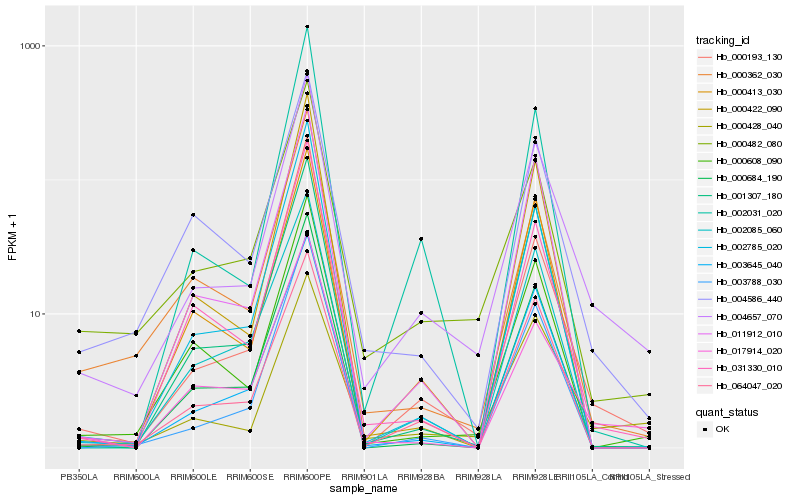
Hb_004657_070 |
0.0 |
- |
- |
PREDICTED: protein IRX15-LIKE [Jatropha curcas] |
| 2 |
Hb_000428_040 |
0.0985253139 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 3 |
Hb_000482_080 |
0.1050129195 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 4 |
Hb_000193_130 |
0.1225118429 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000608_090 |
0.1229750593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_031330_010 |
0.1243387492 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 7 |
Hb_004586_440 |
0.124818032 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 8 |
Hb_017914_020 |
0.1248728885 |
- |
- |
- |
| 9 |
Hb_064047_020 |
0.1286676538 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 10 |
Hb_003645_040 |
0.1287344245 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 11 |
Hb_002031_020 |
0.1296103661 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 12 |
Hb_003788_030 |
0.1296576273 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002785_020 |
0.1320485338 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 14 |
Hb_011912_010 |
0.1329178467 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000362_030 |
0.1342155288 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 16 |
Hb_000684_190 |
0.1342770162 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000422_090 |
0.1346753292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002085_060 |
0.1349248262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000413_030 |
0.1362381131 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 20 |
Hb_001307_180 |
0.1367226555 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |