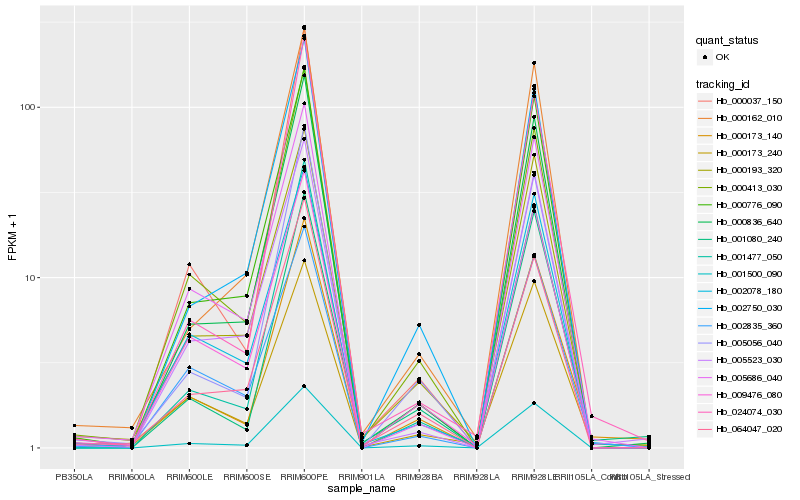
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_140 |
0.0 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 2 |
Hb_001500_090 |
0.0756341922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001080_240 |
0.0798175389 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 4 |
Hb_005056_040 |
0.0850714822 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 5 |
Hb_000173_240 |
0.0855964939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001477_050 |
0.0890859846 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000193_320 |
0.0906754154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000836_640 |
0.0936394254 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 9 |
Hb_002750_030 |
0.0966015687 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 10 |
Hb_009476_080 |
0.0985963158 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 11 |
Hb_000413_030 |
0.0994345668 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 12 |
Hb_002078_180 |
0.101338108 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 13 |
Hb_005686_040 |
0.1015442113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005523_030 |
0.1026901859 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_000776_090 |
0.1037351734 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 16 |
Hb_000162_010 |
0.1071793775 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 17 |
Hb_002835_360 |
0.107617825 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 18 |
Hb_024074_030 |
0.108039337 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 19 |
Hb_064047_020 |
0.1084227463 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 20 |
Hb_000037_150 |
0.1091850342 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |