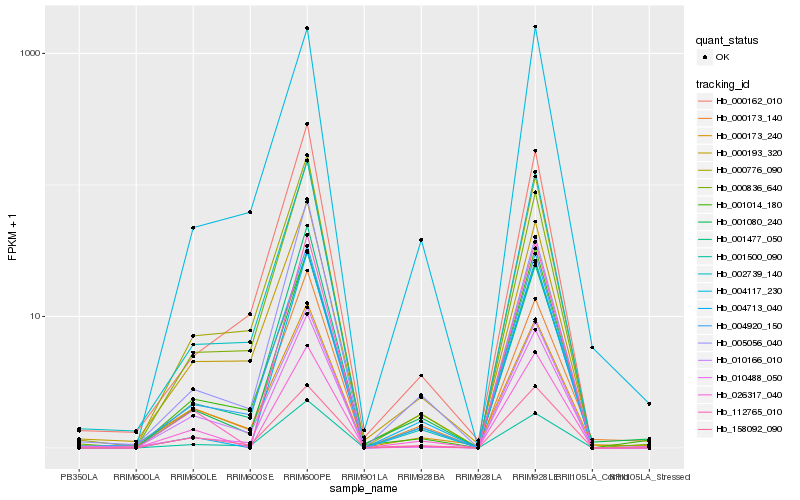
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_240 |
0.0 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 2 |
Hb_004920_150 |
0.0786445009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000173_140 |
0.0798175389 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 4 |
Hb_001500_090 |
0.088376622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004117_230 |
0.0906104525 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 6 |
Hb_010488_050 |
0.0951412261 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 7 |
Hb_004713_040 |
0.0957055475 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 8 |
Hb_026317_040 |
0.0968320903 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 9 |
Hb_000193_320 |
0.0994787916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001477_050 |
0.1001356856 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001014_180 |
0.1009076748 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 12 |
Hb_158092_090 |
0.1024677701 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000162_010 |
0.1032585321 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 14 |
Hb_000776_090 |
0.1042241644 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 15 |
Hb_005056_040 |
0.1043559565 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 16 |
Hb_000836_640 |
0.1044213552 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 17 |
Hb_002739_140 |
0.1065403014 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 18 |
Hb_000173_240 |
0.1068178772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010166_010 |
0.1071785322 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_112765_010 |
0.1092741421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |