| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
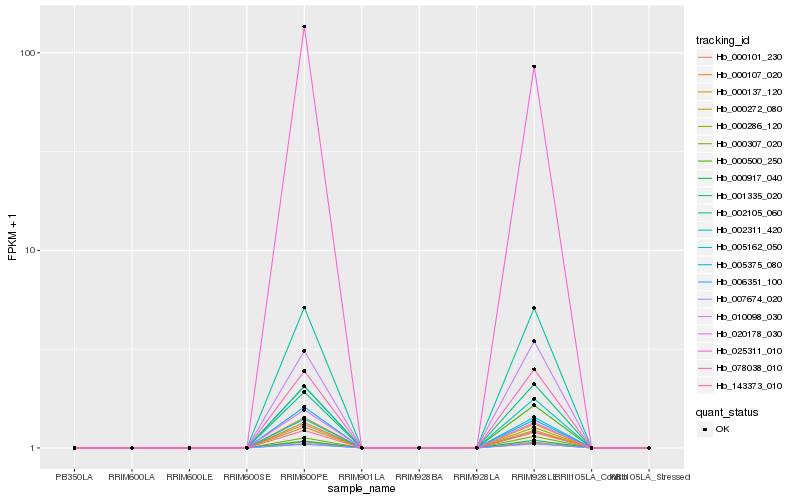
Hb_143373_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 2 |
Hb_000107_020 |
0.0136688727 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 3 |
Hb_007674_020 |
0.0220646125 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 4 |
Hb_000272_080 |
0.0249716594 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 5 |
Hb_002105_060 |
0.0263785136 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 6 |
Hb_000137_120 |
0.0301362193 |
- |
- |
hypothetical protein JCGZ_15021 [Jatropha curcas] |
| 7 |
Hb_078038_010 |
0.0348262548 |
- |
- |
hypothetical protein CISIN_1g038197mg [Citrus sinensis] |
| 8 |
Hb_002311_420 |
0.0372919597 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 9 |
Hb_005162_050 |
0.0435821826 |
- |
- |
hypothetical protein B456_009G352100 [Gossypium raimondii] |
| 10 |
Hb_000917_040 |
0.052507035 |
- |
- |
PREDICTED: uncharacterized protein LOC105639996 [Jatropha curcas] |
| 11 |
Hb_000307_020 |
0.0595415617 |
- |
- |
PREDICTED: uncharacterized protein LOC100255757 [Vitis vinifera] |
| 12 |
Hb_000286_120 |
0.0599309244 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 13 |
Hb_000101_230 |
0.0608047155 |
- |
- |
hypothetical protein CICLE_v10030840mg [Citrus clementina] |
| 14 |
Hb_010098_030 |
0.0623654398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001335_020 |
0.0668736589 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 16 |
Hb_005375_080 |
0.0681445541 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 17 |
Hb_006351_100 |
0.069297482 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_025311_010 |
0.0706893377 |
- |
- |
- |
| 19 |
Hb_020178_030 |
0.0728631206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000500_250 |
0.0748072306 |
- |
- |
hypothetical protein CISIN_1g006096mg [Citrus sinensis] |