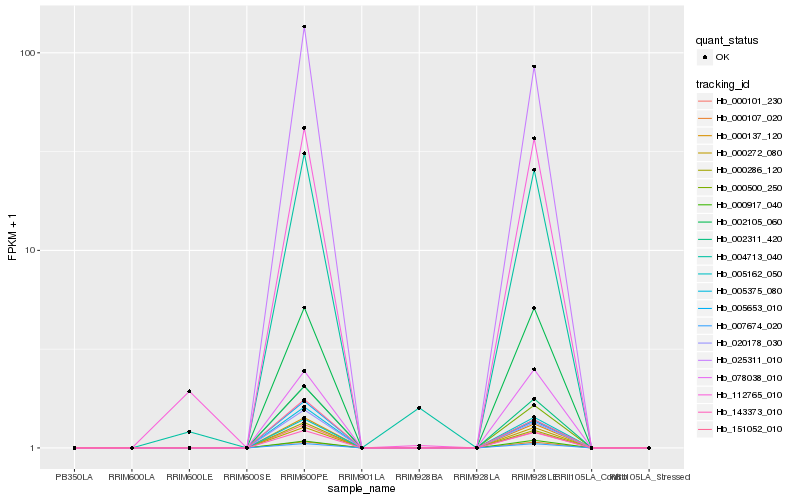
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000137_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_15021 [Jatropha curcas] |
| 2 |
Hb_002311_420 |
0.0071586083 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 3 |
Hb_000286_120 |
0.0298140741 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 4 |
Hb_143373_010 |
0.0301362193 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 5 |
Hb_000101_230 |
0.0306887317 |
- |
- |
hypothetical protein CICLE_v10030840mg [Citrus clementina] |
| 6 |
Hb_005375_080 |
0.0380365329 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 7 |
Hb_025311_010 |
0.0405843645 |
- |
- |
- |
| 8 |
Hb_020178_030 |
0.042760869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000107_020 |
0.0437987607 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 10 |
Hb_000500_250 |
0.0447075055 |
- |
- |
hypothetical protein CISIN_1g006096mg [Citrus sinensis] |
| 11 |
Hb_007674_020 |
0.0521886781 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 12 |
Hb_000272_080 |
0.0550933675 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 13 |
Hb_002105_060 |
0.0564990179 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 14 |
Hb_078038_010 |
0.064938668 |
- |
- |
hypothetical protein CISIN_1g038197mg [Citrus sinensis] |
| 15 |
Hb_005162_050 |
0.0736846522 |
- |
- |
hypothetical protein B456_009G352100 [Gossypium raimondii] |
| 16 |
Hb_005653_010 |
0.0810841144 |
- |
- |
CEN-like protein 2 [Jatropha curcas] |
| 17 |
Hb_151052_010 |
0.0811733609 |
- |
- |
- |
| 18 |
Hb_000917_040 |
0.08259774 |
- |
- |
PREDICTED: uncharacterized protein LOC105639996 [Jatropha curcas] |
| 19 |
Hb_112765_010 |
0.0850455633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004713_040 |
0.0868760696 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |