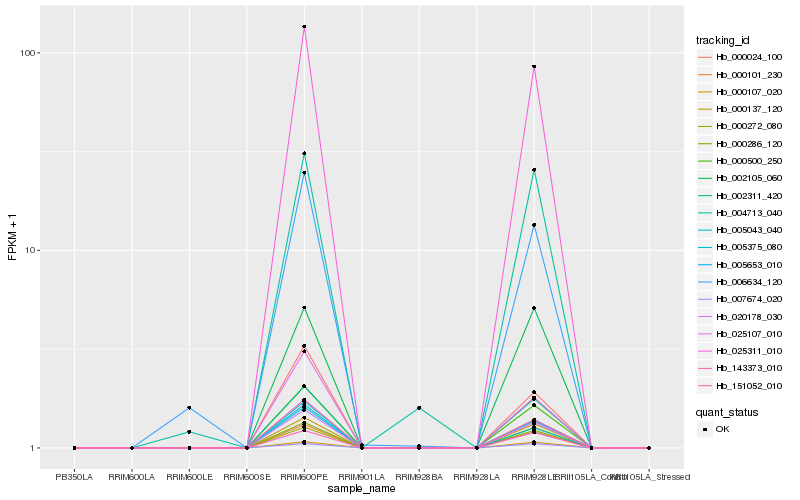
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005375_080 |
0.0 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 2 |
Hb_025311_010 |
0.0025493061 |
- |
- |
- |
| 3 |
Hb_020178_030 |
0.0047272208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000500_250 |
0.0066752365 |
- |
- |
hypothetical protein CISIN_1g006096mg [Citrus sinensis] |
| 5 |
Hb_000101_230 |
0.0073509973 |
- |
- |
hypothetical protein CICLE_v10030840mg [Citrus clementina] |
| 6 |
Hb_000286_120 |
0.0082259315 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 7 |
Hb_002311_420 |
0.0308810156 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 8 |
Hb_000137_120 |
0.0380365329 |
- |
- |
hypothetical protein JCGZ_15021 [Jatropha curcas] |
| 9 |
Hb_005653_010 |
0.043098725 |
- |
- |
CEN-like protein 2 [Jatropha curcas] |
| 10 |
Hb_151052_010 |
0.0431881373 |
- |
- |
- |
| 11 |
Hb_143373_010 |
0.0681445541 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 12 |
Hb_000107_020 |
0.0817863926 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 13 |
Hb_007674_020 |
0.0901611906 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 14 |
Hb_000024_100 |
0.0915895773 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 15 |
Hb_005043_040 |
0.091723988 |
- |
- |
hypothetical protein POPTR_0004s17180g [Populus trichocarpa] |
| 16 |
Hb_000272_080 |
0.0930602217 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 17 |
Hb_002105_060 |
0.0944630559 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 18 |
Hb_025107_010 |
0.0992334679 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like [Sesamum indicum] |
| 19 |
Hb_004713_040 |
0.1010926892 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 20 |
Hb_006634_120 |
0.1018917036 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |