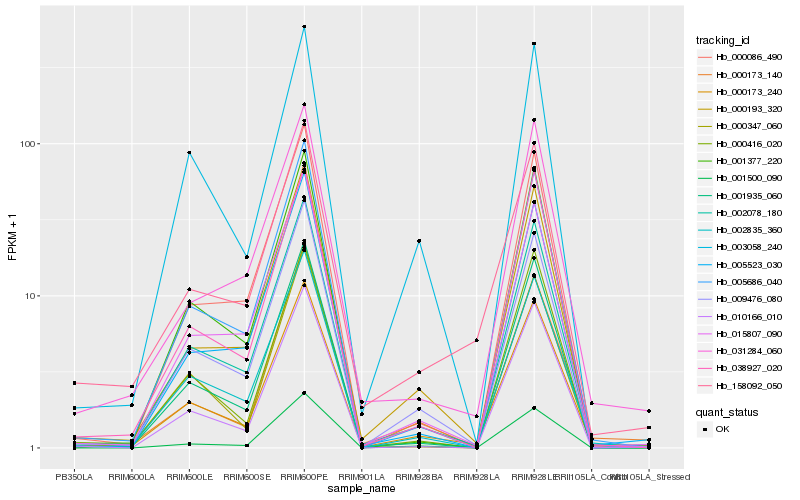
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002078_180 |
0.0543502286 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 3 |
Hb_000193_320 |
0.0657825618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009476_080 |
0.0697391989 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 5 |
Hb_002835_360 |
0.0708932363 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 6 |
Hb_001935_060 |
0.0765821168 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 7 |
Hb_005686_040 |
0.0777108112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010166_010 |
0.0812644372 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 9 |
Hb_038927_020 |
0.0827887316 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 10 |
Hb_000416_020 |
0.0843299956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000173_140 |
0.0855964939 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 12 |
Hb_003058_240 |
0.0859221014 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 13 |
Hb_031284_060 |
0.0864877812 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_015807_090 |
0.0871778651 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_001377_220 |
0.0874497698 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 16 |
Hb_000347_060 |
0.0875245651 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |
| 17 |
Hb_001500_090 |
0.0889194815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_158092_050 |
0.0894813452 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 19 |
Hb_000086_490 |
0.0909108157 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 20 |
Hb_005523_030 |
0.0909499925 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |