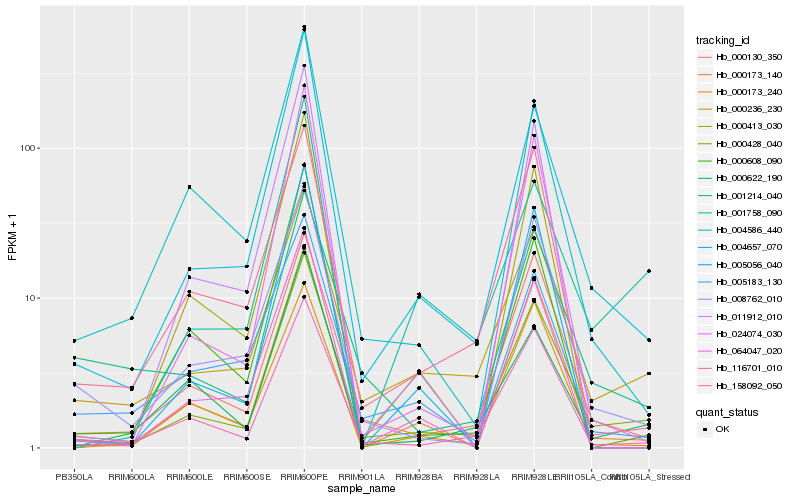
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000428_040 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 2 |
Hb_004657_070 |
0.0985253139 |
- |
- |
PREDICTED: protein IRX15-LIKE [Jatropha curcas] |
| 3 |
Hb_116701_010 |
0.1062510811 |
- |
- |
hypothetical protein POPTR_0005s19410g [Populus trichocarpa] |
| 4 |
Hb_000236_230 |
0.1073923353 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000130_350 |
0.1205333683 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 6 |
Hb_008762_010 |
0.1289722634 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_000173_140 |
0.1299013326 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 8 |
Hb_024074_030 |
0.1492800226 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 9 |
Hb_000622_190 |
0.1517105463 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 10 |
Hb_000173_240 |
0.1528021469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000413_030 |
0.1551928181 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 12 |
Hb_158092_050 |
0.1553672985 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 13 |
Hb_064047_020 |
0.1554904284 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 14 |
Hb_000608_090 |
0.1560886951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005056_040 |
0.1565638093 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 16 |
Hb_005183_130 |
0.156683258 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 17 |
Hb_004586_440 |
0.1569938078 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 18 |
Hb_001758_090 |
0.1570289124 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 19 |
Hb_011912_010 |
0.1576607285 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001214_040 |
0.158487287 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |