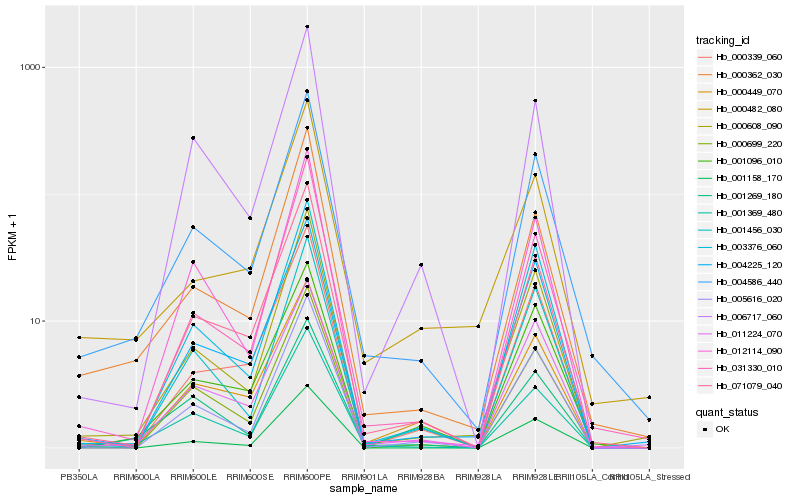
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_440 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_000608_090 |
0.0810325514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001369_480 |
0.0811503751 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 4 |
Hb_000362_030 |
0.0844230875 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_071079_040 |
0.0879157827 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 6 |
Hb_031330_010 |
0.0958888088 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 7 |
Hb_011224_070 |
0.1023636169 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 8 |
Hb_003376_060 |
0.1052371816 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005616_020 |
0.1057937027 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_001456_030 |
0.1067076682 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 11 |
Hb_004225_120 |
0.1082200499 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 12 |
Hb_000339_060 |
0.1088792941 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001096_010 |
0.1091516881 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 14 |
Hb_012114_090 |
0.1092060971 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 15 |
Hb_001269_180 |
0.1096319562 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000449_070 |
0.1108911186 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000482_080 |
0.1116293171 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 18 |
Hb_000699_220 |
0.1119117956 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 19 |
Hb_006717_060 |
0.1125773707 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 20 |
Hb_001158_170 |
0.1139972029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |