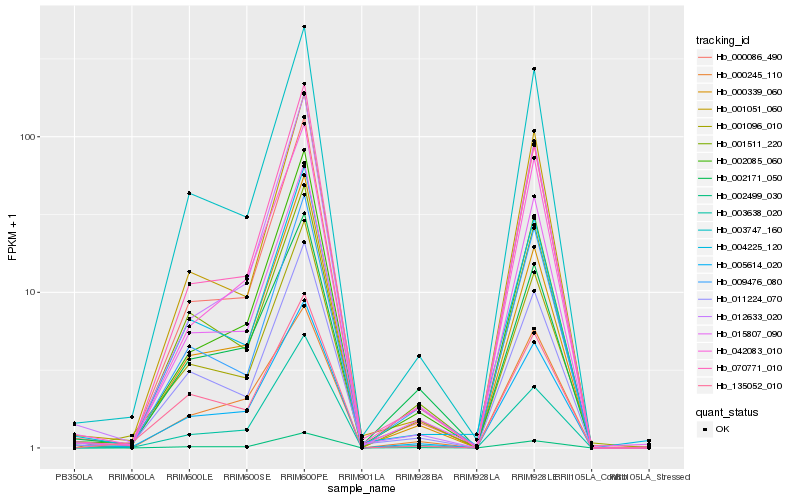
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005614_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003747_160 |
0.0591361847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002499_030 |
0.0601871553 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_011224_070 |
0.0659464585 |
- |
- |
PREDICTED: solute carrier family 35 member E3 [Jatropha curcas] |
| 5 |
Hb_015807_090 |
0.0687957554 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 6 |
Hb_004225_120 |
0.0706753741 |
- |
- |
PREDICTED: uncharacterized protein LOC105628059 [Jatropha curcas] |
| 7 |
Hb_042083_010 |
0.0720332765 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 8 |
Hb_001096_010 |
0.073425949 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 9 |
Hb_070771_010 |
0.0752156973 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 10 |
Hb_002085_060 |
0.0759243914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000339_060 |
0.0765968395 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003638_020 |
0.0800249019 |
- |
- |
PREDICTED: uncharacterized protein LOC105645807 [Jatropha curcas] |
| 13 |
Hb_000245_110 |
0.0807922644 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 14 |
Hb_001511_220 |
0.0808207474 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 15 |
Hb_002171_050 |
0.0833008822 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 16 |
Hb_135052_010 |
0.0839650845 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_001051_060 |
0.0852010614 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 18 |
Hb_009476_080 |
0.0871861325 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 19 |
Hb_012633_020 |
0.0886319386 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_000086_490 |
0.0900239507 |
- |
- |
cellulose synthase [Populus tomentosa] |