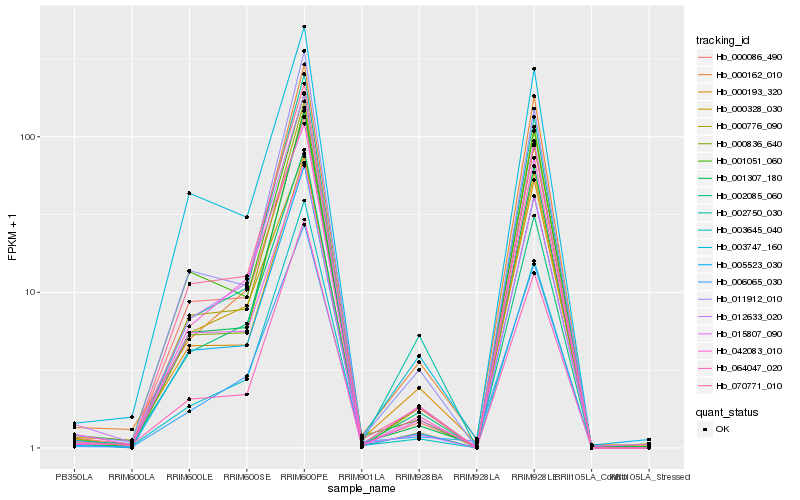
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006065_030 |
0.0 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_042083_010 |
0.0570850316 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 3 |
Hb_012633_020 |
0.0619821427 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_015807_090 |
0.0741150594 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 5 |
Hb_002750_030 |
0.0745617431 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 6 |
Hb_005523_030 |
0.0753505668 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 7 |
Hb_000086_490 |
0.0761149318 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 8 |
Hb_001307_180 |
0.0799124911 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 9 |
Hb_000836_640 |
0.080510789 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 10 |
Hb_003645_040 |
0.0820498581 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 11 |
Hb_000162_010 |
0.0827932253 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 12 |
Hb_070771_010 |
0.0837090202 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 13 |
Hb_001051_060 |
0.084640405 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 14 |
Hb_000776_090 |
0.0847856468 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 15 |
Hb_000193_320 |
0.0850873067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_064047_020 |
0.086397883 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 17 |
Hb_002085_060 |
0.0887863993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003747_160 |
0.091209684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011912_010 |
0.091220762 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000328_030 |
0.092003106 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |