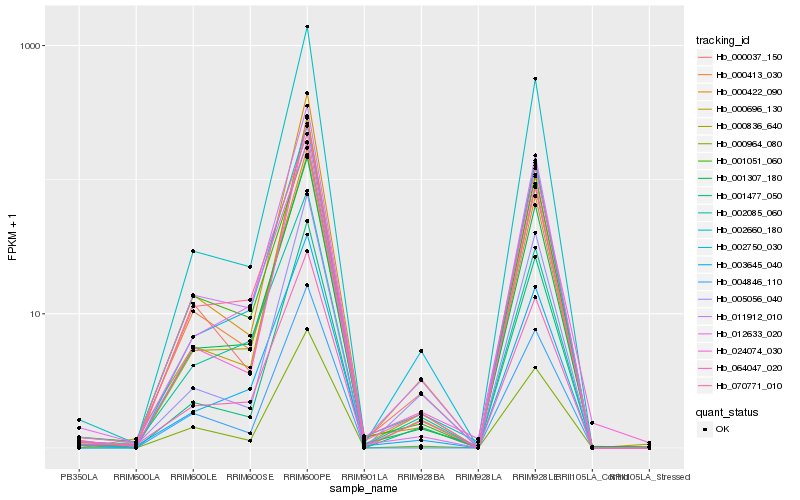
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011912_010 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001307_180 |
0.0283101239 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 3 |
Hb_000413_030 |
0.0401312083 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 4 |
Hb_070771_010 |
0.050489274 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 5 |
Hb_000037_150 |
0.0510507316 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 6 |
Hb_003645_040 |
0.0534484743 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 7 |
Hb_064047_020 |
0.0566088099 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 8 |
Hb_002660_180 |
0.0573746545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004846_110 |
0.0611834099 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 10 |
Hb_000964_080 |
0.0613429531 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 11 |
Hb_000836_640 |
0.06275812 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 12 |
Hb_012633_020 |
0.0633665785 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_002750_030 |
0.0642426939 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 14 |
Hb_001477_050 |
0.0659936688 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002085_060 |
0.0686088782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000696_130 |
0.0719627447 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000422_090 |
0.0729125928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_024074_030 |
0.0735398163 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 19 |
Hb_005056_040 |
0.074622033 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 20 |
Hb_001051_060 |
0.0752889308 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |