| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
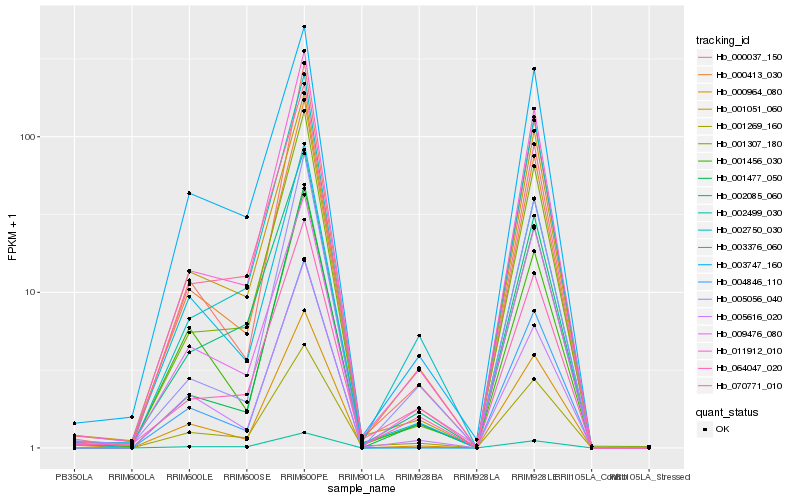
Hb_000413_030 |
0.0 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 2 |
Hb_011912_010 |
0.0401312083 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_064047_020 |
0.0497042172 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 4 |
Hb_000964_080 |
0.0515592145 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 5 |
Hb_003376_060 |
0.0571228838 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000037_150 |
0.0613716371 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 7 |
Hb_001307_180 |
0.0629739538 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 8 |
Hb_070771_010 |
0.0648417136 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 9 |
Hb_001269_160 |
0.0652163059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005616_020 |
0.0662974681 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_004846_110 |
0.073099321 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 12 |
Hb_001051_060 |
0.0743956148 |
- |
- |
UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, putative [Ricinus communis] |
| 13 |
Hb_001456_030 |
0.0749967246 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 14 |
Hb_003747_160 |
0.0753397873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002499_030 |
0.0756984619 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_002750_030 |
0.0764583949 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 17 |
Hb_005056_040 |
0.0769023605 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 18 |
Hb_009476_080 |
0.0777815723 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 19 |
Hb_002085_060 |
0.0786773388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001477_050 |
0.0805795786 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |